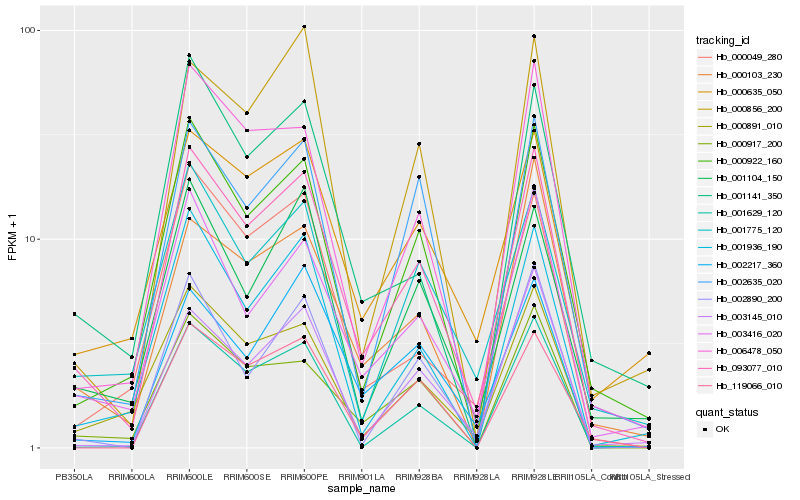
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_093077_010 |
0.0 |
transcription factor |
TF Family: G2-like |
PREDICTED: two-component response regulator-like APRR2 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000922_160 |
0.0999097834 |
rubber biosynthesis |
Gene Name: 1-deoxy-D-xylulose 5-phosphate reductoisomerase |
putative 1-deoxy-D-xylulose 5-phosphate reductoisomerase [Hevea brasiliensis] |
| 3 |
Hb_003416_020 |
0.1099039559 |
- |
- |
pentatricopeptide repeat-containing family protein [Populus trichocarpa] |
| 4 |
Hb_006478_050 |
0.1140953779 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000891_010 |
0.1169949945 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000917_200 |
0.1222141003 |
- |
- |
hypothetical protein PRUPE_ppa010555mg [Prunus persica] |
| 7 |
Hb_001775_120 |
0.1258051568 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 12 [Jatropha curcas] |
| 8 |
Hb_001936_190 |
0.1261244202 |
- |
- |
hypothetical protein JCGZ_17780 [Jatropha curcas] |
| 9 |
Hb_002635_020 |
0.1264016346 |
- |
- |
hypothetical protein POPTR_0018s09790g [Populus trichocarpa] |
| 10 |
Hb_000635_050 |
0.1275705919 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At1g67480 [Jatropha curcas] |
| 11 |
Hb_001141_350 |
0.1288161145 |
- |
- |
hypothetical protein JCGZ_02174 [Jatropha curcas] |
| 12 |
Hb_001104_150 |
0.1290279245 |
- |
- |
PREDICTED: protein ROS1 [Jatropha curcas] |
| 13 |
Hb_003145_010 |
0.1315791476 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein At1g59780 [Jatropha curcas] |
| 14 |
Hb_001629_120 |
0.133553751 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: probable disease resistance protein At4g27220 isoform X2 [Populus euphratica] |
| 15 |
Hb_119066_010 |
0.134359414 |
- |
- |
PREDICTED: uncharacterized protein LOC105641552 [Jatropha curcas] |
| 16 |
Hb_000049_280 |
0.1349436062 |
- |
- |
PREDICTED: ribokinase-like isoform X4 [Jatropha curcas] |
| 17 |
Hb_000103_230 |
0.1368387038 |
- |
- |
PREDICTED: chlorophyllase-2, chloroplastic [Jatropha curcas] |
| 18 |
Hb_002890_200 |
0.1369669369 |
- |
- |
PREDICTED: methionine S-methyltransferase [Jatropha curcas] |
| 19 |
Hb_002217_360 |
0.1372165872 |
- |
- |
PREDICTED: nuclear poly(A) polymerase 3 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000856_200 |
0.1375367157 |
- |
- |
ATPase 4, plasma membrane-type -like protein [Gossypium arboreum] |