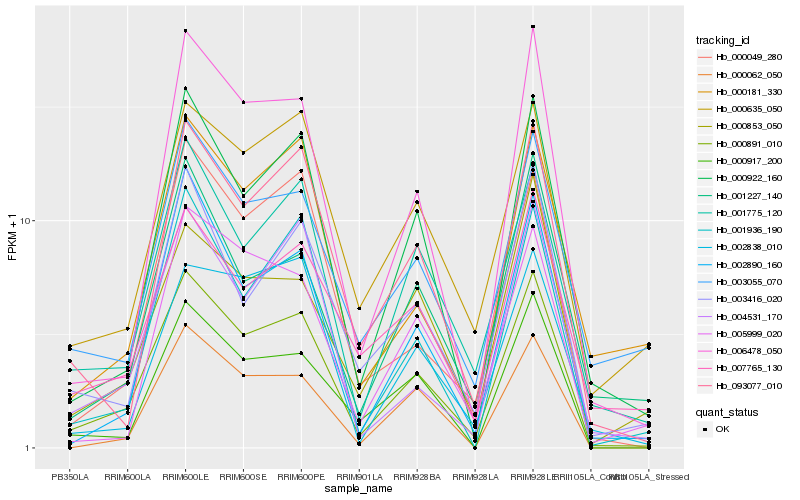
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000891_010 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000922_160 |
0.1048630435 |
rubber biosynthesis |
Gene Name: 1-deoxy-D-xylulose 5-phosphate reductoisomerase |
putative 1-deoxy-D-xylulose 5-phosphate reductoisomerase [Hevea brasiliensis] |
| 3 |
Hb_006478_050 |
0.1082690562 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1 isoform X1 [Jatropha curcas] |
| 4 |
Hb_001775_120 |
0.111618853 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 12 [Jatropha curcas] |
| 5 |
Hb_093077_010 |
0.1169949945 |
transcription factor |
TF Family: G2-like |
PREDICTED: two-component response regulator-like APRR2 isoform X1 [Jatropha curcas] |
| 6 |
Hb_002890_160 |
0.1217244854 |
- |
- |
PREDICTED: protein root UVB sensitive 6 isoform X1 [Jatropha curcas] |
| 7 |
Hb_001936_190 |
0.1240348964 |
- |
- |
hypothetical protein JCGZ_17780 [Jatropha curcas] |
| 8 |
Hb_000917_200 |
0.1267215123 |
- |
- |
hypothetical protein PRUPE_ppa010555mg [Prunus persica] |
| 9 |
Hb_003416_020 |
0.1275692662 |
- |
- |
pentatricopeptide repeat-containing family protein [Populus trichocarpa] |
| 10 |
Hb_004531_170 |
0.1301041225 |
- |
- |
PREDICTED: probable inactive nicotinamidase At3g16190 [Jatropha curcas] |
| 11 |
Hb_000049_280 |
0.1302750785 |
- |
- |
PREDICTED: ribokinase-like isoform X4 [Jatropha curcas] |
| 12 |
Hb_000635_050 |
0.13331944 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At1g67480 [Jatropha curcas] |
| 13 |
Hb_007765_130 |
0.133964805 |
- |
- |
PREDICTED: UPF0503 protein At3g09070, chloroplastic [Jatropha curcas] |
| 14 |
Hb_000062_050 |
0.1352977947 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 15 |
Hb_003055_070 |
0.1355125187 |
- |
- |
PREDICTED: katanin p80 WD40 repeat-containing subunit B1 homolog isoform X1 [Jatropha curcas] |
| 16 |
Hb_001227_140 |
0.1396283483 |
- |
- |
PREDICTED: protein ROS1 [Jatropha curcas] |
| 17 |
Hb_000853_050 |
0.1428676185 |
- |
- |
PREDICTED: putative GTP diphosphokinase RSH1, chloroplastic isoform X1 [Jatropha curcas] |
| 18 |
Hb_002838_010 |
0.1445895313 |
- |
- |
AGPase large subunit protein [Hevea brasiliensis] |
| 19 |
Hb_005999_020 |
0.1461122437 |
transcription factor |
TF Family: C2H2 |
PREDICTED: protein SENSITIVE TO PROTON RHIZOTOXICITY 1-like [Jatropha curcas] |
| 20 |
Hb_000181_330 |
0.1462727779 |
- |
- |
ubiquitin ligase protein cop1, putative [Ricinus communis] |