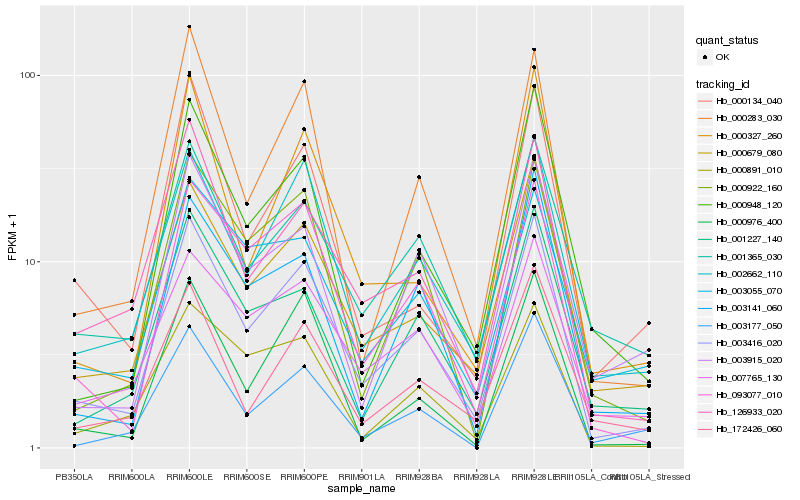
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003416_020 |
0.0 |
- |
- |
pentatricopeptide repeat-containing family protein [Populus trichocarpa] |
| 2 |
Hb_000679_080 |
0.0936988104 |
- |
- |
PREDICTED: lactation elevated protein 1 isoform X1 [Jatropha curcas] |
| 3 |
Hb_002662_110 |
0.0985670369 |
- |
- |
Kinesin heavy chain, putative [Ricinus communis] |
| 4 |
Hb_126933_020 |
0.1044487492 |
- |
- |
PREDICTED: uncharacterized protein LOC105638525 [Jatropha curcas] |
| 5 |
Hb_093077_010 |
0.1099039559 |
transcription factor |
TF Family: G2-like |
PREDICTED: two-component response regulator-like APRR2 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000283_030 |
0.1102726191 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA16-like [Populus euphratica] |
| 7 |
Hb_007765_130 |
0.1152503423 |
- |
- |
PREDICTED: UPF0503 protein At3g09070, chloroplastic [Jatropha curcas] |
| 8 |
Hb_001365_030 |
0.1169351711 |
- |
- |
PREDICTED: uncharacterized protein LOC102609547 [Citrus sinensis] |
| 9 |
Hb_000922_160 |
0.1191215681 |
rubber biosynthesis |
Gene Name: 1-deoxy-D-xylulose 5-phosphate reductoisomerase |
putative 1-deoxy-D-xylulose 5-phosphate reductoisomerase [Hevea brasiliensis] |
| 10 |
Hb_000948_120 |
0.1192135717 |
- |
- |
PREDICTED: uncharacterized protein LOC105634688 isoform X1 [Jatropha curcas] |
| 11 |
Hb_001227_140 |
0.1208074673 |
- |
- |
PREDICTED: protein ROS1 [Jatropha curcas] |
| 12 |
Hb_003141_060 |
0.1274709673 |
transcription factor |
TF Family: TRAF |
PREDICTED: ETO1-like protein 1 [Jatropha curcas] |
| 13 |
Hb_000891_010 |
0.1275692662 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000327_260 |
0.1294178885 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_003915_020 |
0.1299036642 |
- |
- |
PREDICTED: outer envelope pore protein 37, chloroplastic [Jatropha curcas] |
| 16 |
Hb_172426_060 |
0.131905197 |
- |
- |
PREDICTED: protein BREAST CANCER SUSCEPTIBILITY 1 homolog [Jatropha curcas] |
| 17 |
Hb_000976_400 |
0.1332275352 |
- |
- |
synaptotagmin, putative [Ricinus communis] |
| 18 |
Hb_003055_070 |
0.1340820039 |
- |
- |
PREDICTED: katanin p80 WD40 repeat-containing subunit B1 homolog isoform X1 [Jatropha curcas] |
| 19 |
Hb_003177_050 |
0.1342274567 |
- |
- |
PREDICTED: cytokinin riboside 5'-monophosphate phosphoribohydrolase LOG1-like [Populus euphratica] |
| 20 |
Hb_000134_040 |
0.1347602404 |
- |
- |
PREDICTED: 30S ribosomal protein 2, chloroplastic [Jatropha curcas] |