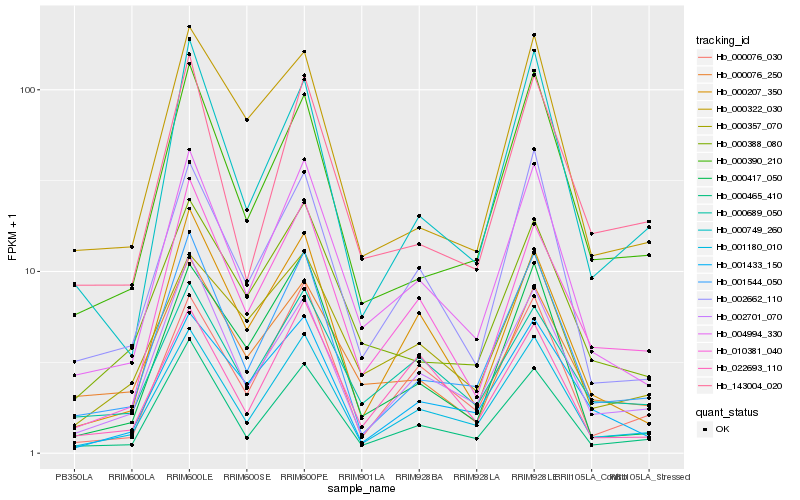
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004994_330 |
0.0 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 2 |
Hb_001180_010 |
0.0809457559 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 9 [Jatropha curcas] |
| 3 |
Hb_002662_110 |
0.0864504101 |
- |
- |
Kinesin heavy chain, putative [Ricinus communis] |
| 4 |
Hb_000465_410 |
0.0978707284 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_001544_050 |
0.0981885478 |
- |
- |
PREDICTED: U-box domain-containing protein 11-like [Jatropha curcas] |
| 6 |
Hb_000417_050 |
0.1037680496 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase MRH1 [Jatropha curcas] |
| 7 |
Hb_000207_350 |
0.105078976 |
- |
- |
PREDICTED: embryogenesis-associated protein EMB8 [Jatropha curcas] |
| 8 |
Hb_000076_030 |
0.1123752202 |
- |
- |
PREDICTED: microtubule-associated protein TORTIFOLIA1 isoform X1 [Jatropha curcas] |
| 9 |
Hb_002701_070 |
0.1136771366 |
- |
- |
PREDICTED: uncharacterized protein LOC105646760 [Jatropha curcas] |
| 10 |
Hb_000388_080 |
0.1146061641 |
- |
- |
PREDICTED: serine/threonine-protein kinase CDL1 [Jatropha curcas] |
| 11 |
Hb_010381_040 |
0.1172134914 |
- |
- |
beta-galactosidase, putative [Ricinus communis] |
| 12 |
Hb_000390_210 |
0.119469349 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 13 |
Hb_001433_150 |
0.1205566591 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000357_070 |
0.1215205664 |
- |
- |
- |
| 15 |
Hb_000689_050 |
0.121935569 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_022693_110 |
0.1224461902 |
- |
- |
phosphate transporter [Manihot esculenta] |
| 17 |
Hb_000076_250 |
0.1261664785 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000322_030 |
0.1267303217 |
- |
- |
PREDICTED: uncharacterized protein LOC105637911 [Jatropha curcas] |
| 19 |
Hb_143004_020 |
0.1267847894 |
- |
- |
PREDICTED: phosphopantothenate--cysteine ligase 2-like [Jatropha curcas] |
| 20 |
Hb_000749_260 |
0.1270582248 |
- |
- |
PREDICTED: protochlorophyllide reductase, chloroplastic [Jatropha curcas] |