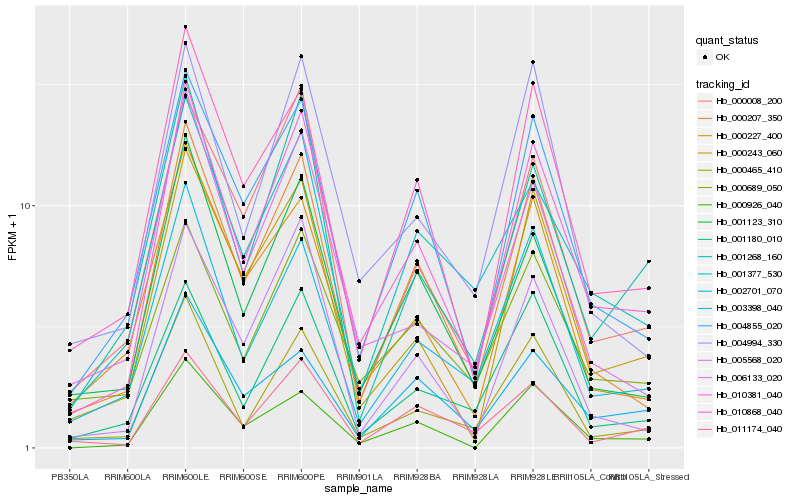
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010381_040 |
0.0 |
- |
- |
beta-galactosidase, putative [Ricinus communis] |
| 2 |
Hb_000207_350 |
0.0916611777 |
- |
- |
PREDICTED: embryogenesis-associated protein EMB8 [Jatropha curcas] |
| 3 |
Hb_004855_020 |
0.0958271447 |
- |
- |
hypothetical protein POPTR_0006s10010g [Populus trichocarpa] |
| 4 |
Hb_010868_040 |
0.098199687 |
- |
- |
PREDICTED: reticulon-like protein B21 isoform X1 [Populus euphratica] |
| 5 |
Hb_000243_060 |
0.1079018313 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 5-like [Jatropha curcas] |
| 6 |
Hb_001123_310 |
0.1098511085 |
- |
- |
PREDICTED: coiled-coil domain-containing protein SCD2 [Jatropha curcas] |
| 7 |
Hb_002701_070 |
0.1116749971 |
- |
- |
PREDICTED: uncharacterized protein LOC105646760 [Jatropha curcas] |
| 8 |
Hb_000465_410 |
0.1132506548 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_004994_330 |
0.1172134914 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 10 |
Hb_000926_040 |
0.1178331645 |
- |
- |
PREDICTED: DNA mismatch repair protein MSH5 [Jatropha curcas] |
| 11 |
Hb_000008_200 |
0.1182919369 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 11 [Jatropha curcas] |
| 12 |
Hb_000689_050 |
0.1210471596 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_011174_040 |
0.1211939473 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_001268_160 |
0.1212332493 |
- |
- |
Glycerophosphodiester phosphodiesterase [Medicago truncatula] |
| 15 |
Hb_001377_530 |
0.1217410113 |
- |
- |
NC domain-containing family protein [Populus trichocarpa] |
| 16 |
Hb_001180_010 |
0.1223533396 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 9 [Jatropha curcas] |
| 17 |
Hb_003398_040 |
0.1237909841 |
- |
- |
PREDICTED: actin-related protein 5 isoform X1 [Jatropha curcas] |
| 18 |
Hb_005568_020 |
0.1257263925 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor RAP2-7 isoform X1 [Jatropha curcas] |
| 19 |
Hb_006133_020 |
0.1258909578 |
- |
- |
PREDICTED: DNA (cytosine-5)-methyltransferase DRM2 [Jatropha curcas] |
| 20 |
Hb_000227_400 |
0.1266734823 |
- |
- |
lipoxygenase, putative [Ricinus communis] |