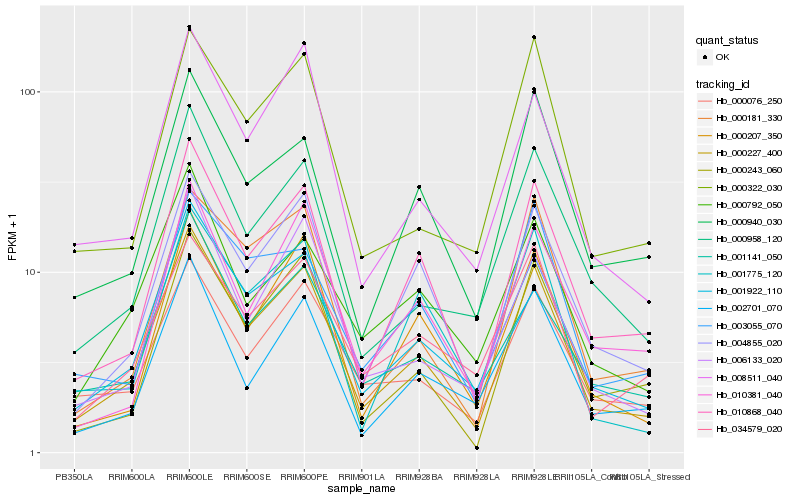
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000227_400 |
0.0 |
- |
- |
lipoxygenase, putative [Ricinus communis] |
| 2 |
Hb_001922_110 |
0.0660183394 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_010868_040 |
0.0795070941 |
- |
- |
PREDICTED: reticulon-like protein B21 isoform X1 [Populus euphratica] |
| 4 |
Hb_004855_020 |
0.0930799273 |
- |
- |
hypothetical protein POPTR_0006s10010g [Populus trichocarpa] |
| 5 |
Hb_000207_350 |
0.1017053252 |
- |
- |
PREDICTED: embryogenesis-associated protein EMB8 [Jatropha curcas] |
| 6 |
Hb_000958_120 |
0.1026746278 |
- |
- |
PREDICTED: uncharacterized protein LOC105646385 [Jatropha curcas] |
| 7 |
Hb_000243_060 |
0.1039609248 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 5-like [Jatropha curcas] |
| 8 |
Hb_008511_040 |
0.1076877951 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 9 |
Hb_006133_020 |
0.1109506783 |
- |
- |
PREDICTED: DNA (cytosine-5)-methyltransferase DRM2 [Jatropha curcas] |
| 10 |
Hb_000076_250 |
0.1116915151 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000322_030 |
0.1154189263 |
- |
- |
PREDICTED: uncharacterized protein LOC105637911 [Jatropha curcas] |
| 12 |
Hb_002701_070 |
0.115590215 |
- |
- |
PREDICTED: uncharacterized protein LOC105646760 [Jatropha curcas] |
| 13 |
Hb_001141_050 |
0.1178585602 |
transcription factor |
TF Family: CPP |
PREDICTED: protein tesmin/TSO1-like CXC 2 isoform X2 [Jatropha curcas] |
| 14 |
Hb_000940_030 |
0.1181932045 |
- |
- |
PREDICTED: triosephosphate isomerase, chloroplastic [Jatropha curcas] |
| 15 |
Hb_000792_050 |
0.1192022539 |
- |
- |
PREDICTED: acetyl-coenzyme A carboxylase carboxyl transferase subunit alpha, chloroplastic isoform X1 [Jatropha curcas] |
| 16 |
Hb_000181_330 |
0.1206821373 |
- |
- |
ubiquitin ligase protein cop1, putative [Ricinus communis] |
| 17 |
Hb_001775_120 |
0.1215915889 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 12 [Jatropha curcas] |
| 18 |
Hb_010381_040 |
0.1266734823 |
- |
- |
beta-galactosidase, putative [Ricinus communis] |
| 19 |
Hb_034579_020 |
0.1269647134 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_003055_070 |
0.1276827324 |
- |
- |
PREDICTED: katanin p80 WD40 repeat-containing subunit B1 homolog isoform X1 [Jatropha curcas] |