| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
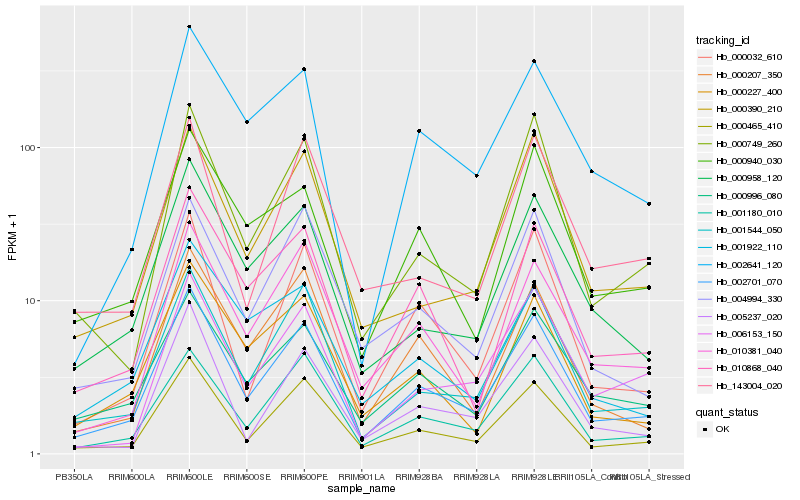
Hb_002701_070 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105646760 [Jatropha curcas] |
| 2 |
Hb_000465_410 |
0.070043217 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_001544_050 |
0.0786679869 |
- |
- |
PREDICTED: U-box domain-containing protein 11-like [Jatropha curcas] |
| 4 |
Hb_001180_010 |
0.0861418532 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 9 [Jatropha curcas] |
| 5 |
Hb_000958_120 |
0.1024900976 |
- |
- |
PREDICTED: uncharacterized protein LOC105646385 [Jatropha curcas] |
| 6 |
Hb_000996_080 |
0.1031306777 |
- |
- |
PREDICTED: intracellular protein transport protein USO1 [Jatropha curcas] |
| 7 |
Hb_010868_040 |
0.1043918826 |
- |
- |
PREDICTED: reticulon-like protein B21 isoform X1 [Populus euphratica] |
| 8 |
Hb_000749_260 |
0.1056399418 |
- |
- |
PREDICTED: protochlorophyllide reductase, chloroplastic [Jatropha curcas] |
| 9 |
Hb_000032_610 |
0.1088481448 |
- |
- |
PREDICTED: 3-oxoacyl-[acyl-carrier-protein] synthase I, chloroplastic-like [Jatropha curcas] |
| 10 |
Hb_000207_350 |
0.1106808668 |
- |
- |
PREDICTED: embryogenesis-associated protein EMB8 [Jatropha curcas] |
| 11 |
Hb_000390_210 |
0.1108666934 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 12 |
Hb_010381_040 |
0.1116749971 |
- |
- |
beta-galactosidase, putative [Ricinus communis] |
| 13 |
Hb_004994_330 |
0.1136771366 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 14 |
Hb_000227_400 |
0.115590215 |
- |
- |
lipoxygenase, putative [Ricinus communis] |
| 15 |
Hb_000940_030 |
0.1169253613 |
- |
- |
PREDICTED: triosephosphate isomerase, chloroplastic [Jatropha curcas] |
| 16 |
Hb_002641_120 |
0.1178875399 |
- |
- |
PREDICTED: calmodulin [Jatropha curcas] |
| 17 |
Hb_005237_020 |
0.121016122 |
- |
- |
Kinesin heavy chain, putative [Ricinus communis] |
| 18 |
Hb_006153_150 |
0.1257283059 |
- |
- |
PREDICTED: uncharacterized protein LOC105645999 [Jatropha curcas] |
| 19 |
Hb_001922_110 |
0.1277267207 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_143004_020 |
0.1286240992 |
- |
- |
PREDICTED: phosphopantothenate--cysteine ligase 2-like [Jatropha curcas] |