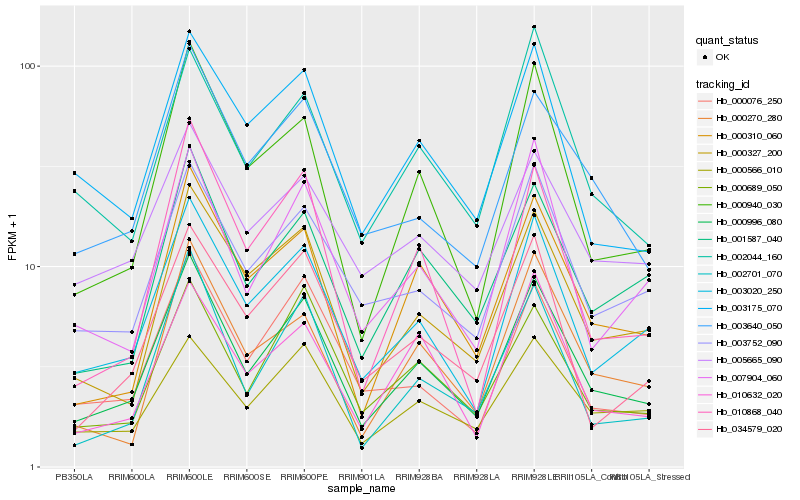
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000996_080 |
0.0 |
- |
- |
PREDICTED: intracellular protein transport protein USO1 [Jatropha curcas] |
| 2 |
Hb_003020_250 |
0.0834164382 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase PP1 isozyme 2 [Jatropha curcas] |
| 3 |
Hb_000940_030 |
0.0944439972 |
- |
- |
PREDICTED: triosephosphate isomerase, chloroplastic [Jatropha curcas] |
| 4 |
Hb_000327_200 |
0.0964100085 |
- |
- |
glutathione-s-transferase omega, putative [Ricinus communis] |
| 5 |
Hb_000310_060 |
0.0970946891 |
- |
- |
hypothetical protein JCGZ_20793 [Jatropha curcas] |
| 6 |
Hb_000689_050 |
0.099833544 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_001587_040 |
0.0999706693 |
- |
- |
PREDICTED: soluble inorganic pyrophosphatase 6, chloroplastic [Jatropha curcas] |
| 8 |
Hb_002701_070 |
0.1031306777 |
- |
- |
PREDICTED: uncharacterized protein LOC105646760 [Jatropha curcas] |
| 9 |
Hb_003640_050 |
0.103471312 |
- |
- |
PREDICTED: protein LUTEIN DEFICIENT 5, chloroplastic isoform X2 [Jatropha curcas] |
| 10 |
Hb_003752_090 |
0.1044420025 |
- |
- |
chitinase, putative [Ricinus communis] |
| 11 |
Hb_000076_250 |
0.1050222759 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000566_010 |
0.1066023639 |
- |
- |
glucose-6-phosphate dehydrogenase [Hevea brasiliensis] |
| 13 |
Hb_007904_060 |
0.1082402558 |
- |
- |
PREDICTED: very-long-chain 3-oxoacyl-CoA reductase 1 [Jatropha curcas] |
| 14 |
Hb_003175_070 |
0.1145555291 |
- |
- |
pyrophosphate-dependent phosphofructokinase, partial [Hevea brasiliensis] |
| 15 |
Hb_000270_280 |
0.1154428155 |
- |
- |
Rhicadhesin receptor precursor, putative [Ricinus communis] |
| 16 |
Hb_010632_020 |
0.1163488148 |
- |
- |
hypothetical protein POPTR_0006s185902g, partial [Populus trichocarpa] |
| 17 |
Hb_005665_090 |
0.1171105198 |
- |
- |
PREDICTED: 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase isoform X2 [Jatropha curcas] |
| 18 |
Hb_034579_020 |
0.1171768708 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_002044_160 |
0.1173786004 |
- |
- |
PREDICTED: glyoxylate/succinic semialdehyde reductase 1 [Jatropha curcas] |
| 20 |
Hb_010868_040 |
0.1180077635 |
- |
- |
PREDICTED: reticulon-like protein B21 isoform X1 [Populus euphratica] |