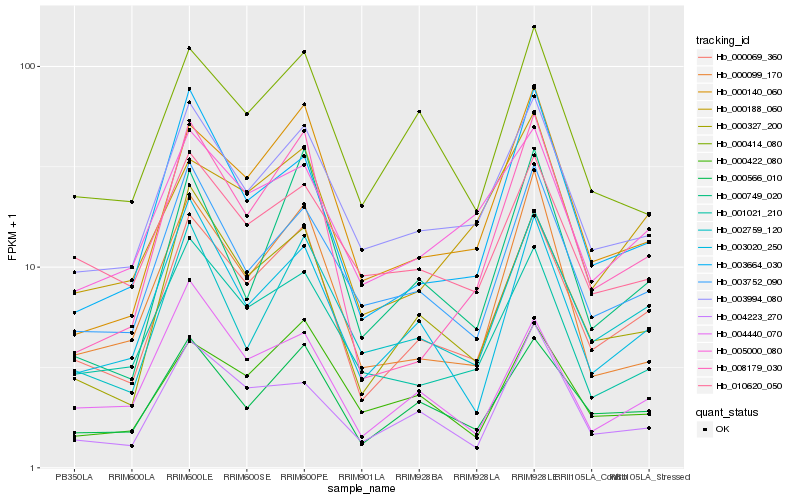
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000069_360 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000566_010 |
0.0870389671 |
- |
- |
glucose-6-phosphate dehydrogenase [Hevea brasiliensis] |
| 3 |
Hb_000327_200 |
0.0870461328 |
- |
- |
glutathione-s-transferase omega, putative [Ricinus communis] |
| 4 |
Hb_003994_080 |
0.0952043408 |
- |
- |
PREDICTED: uncharacterized protein LOC105634384 [Jatropha curcas] |
| 5 |
Hb_000140_060 |
0.1044180387 |
- |
- |
50S ribosomal protein L5, putative [Ricinus communis] |
| 6 |
Hb_003752_090 |
0.1069468508 |
- |
- |
chitinase, putative [Ricinus communis] |
| 7 |
Hb_003020_250 |
0.1111352508 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase PP1 isozyme 2 [Jatropha curcas] |
| 8 |
Hb_002759_120 |
0.113173041 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_001021_210 |
0.1134343761 |
transcription factor |
TF Family: mTERF |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_008179_030 |
0.1211127179 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 8 [Jatropha curcas] |
| 11 |
Hb_000422_080 |
0.1221355044 |
transcription factor |
TF Family: E2F-DP |
PREDICTED: transcription factor E2FB isoform X1 [Jatropha curcas] |
| 12 |
Hb_003664_030 |
0.1222888863 |
- |
- |
PREDICTED: mitochondrial carnitine/acylcarnitine carrier-like protein [Jatropha curcas] |
| 13 |
Hb_000188_060 |
0.1251620171 |
- |
- |
PREDICTED: zinc finger A20 and AN1 domain-containing stress-associated protein 4-like [Jatropha curcas] |
| 14 |
Hb_005000_080 |
0.1258179546 |
- |
- |
PREDICTED: thioredoxin-like 2, chloroplastic [Populus euphratica] |
| 15 |
Hb_004440_070 |
0.1272908477 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g66520 [Jatropha curcas] |
| 16 |
Hb_000414_080 |
0.1280634217 |
- |
- |
PREDICTED: triosephosphate isomerase, chloroplastic [Jatropha curcas] |
| 17 |
Hb_000099_170 |
0.130376562 |
- |
- |
PREDICTED: uncharacterized protein LOC105645733 [Jatropha curcas] |
| 18 |
Hb_000749_020 |
0.1305689652 |
- |
- |
Nucleotide-diphospho-sugar transferases superfamily protein isoform 1 [Theobroma cacao] |
| 19 |
Hb_004223_270 |
0.1306738963 |
- |
- |
PREDICTED: uncharacterized protein LOC105635371 isoform X1 [Jatropha curcas] |
| 20 |
Hb_010620_050 |
0.1309040279 |
- |
- |
PREDICTED: uncharacterized protein LOC105646119 [Jatropha curcas] |