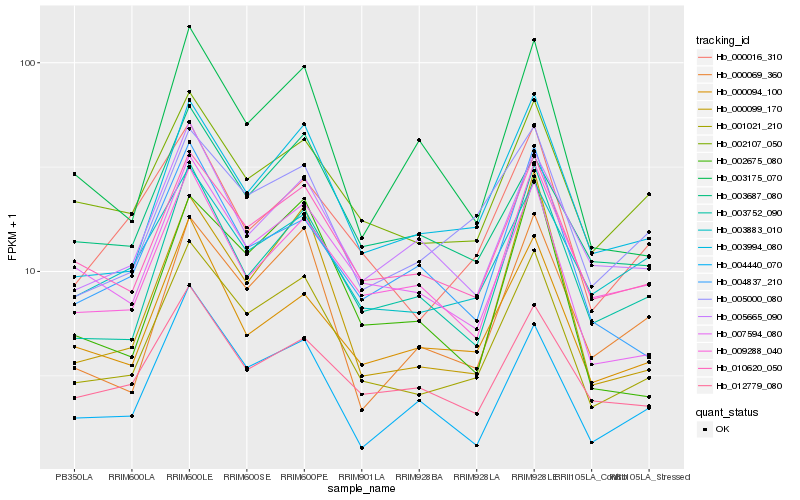
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001021_210 |
0.0 |
transcription factor |
TF Family: mTERF |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_003994_080 |
0.0798487658 |
- |
- |
PREDICTED: uncharacterized protein LOC105634384 [Jatropha curcas] |
| 3 |
Hb_010620_050 |
0.0862529907 |
- |
- |
PREDICTED: uncharacterized protein LOC105646119 [Jatropha curcas] |
| 4 |
Hb_003752_090 |
0.0963148508 |
- |
- |
chitinase, putative [Ricinus communis] |
| 5 |
Hb_005000_080 |
0.1029059956 |
- |
- |
PREDICTED: thioredoxin-like 2, chloroplastic [Populus euphratica] |
| 6 |
Hb_002107_050 |
0.1052157361 |
- |
- |
PREDICTED: prostaglandin E synthase 2 [Jatropha curcas] |
| 7 |
Hb_007594_080 |
0.1054234121 |
- |
- |
PREDICTED: polygalacturonate 4-alpha-galacturonosyltransferase [Jatropha curcas] |
| 8 |
Hb_000094_100 |
0.1095067526 |
- |
- |
PREDICTED: probable methyltransferase PMT28 [Jatropha curcas] |
| 9 |
Hb_012779_080 |
0.1111585489 |
- |
- |
PREDICTED: carbon catabolite repressor protein 4 homolog 3 isoform X3 [Jatropha curcas] |
| 10 |
Hb_000016_310 |
0.1112570984 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 11 |
Hb_000069_360 |
0.1134343761 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_004440_070 |
0.1144911827 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g66520 [Jatropha curcas] |
| 13 |
Hb_000099_170 |
0.1147038842 |
- |
- |
PREDICTED: uncharacterized protein LOC105645733 [Jatropha curcas] |
| 14 |
Hb_005665_090 |
0.115776921 |
- |
- |
PREDICTED: 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase isoform X2 [Jatropha curcas] |
| 15 |
Hb_002675_080 |
0.1171342821 |
- |
- |
PREDICTED: uncharacterized protein LOC105634950 [Jatropha curcas] |
| 16 |
Hb_003175_070 |
0.1172645916 |
- |
- |
pyrophosphate-dependent phosphofructokinase, partial [Hevea brasiliensis] |
| 17 |
Hb_003687_080 |
0.1190559626 |
- |
- |
PREDICTED: casein kinase I-like [Jatropha curcas] |
| 18 |
Hb_009288_040 |
0.1191999592 |
- |
- |
PREDICTED: uncharacterized protein LOC105648677 [Jatropha curcas] |
| 19 |
Hb_004837_210 |
0.1209277559 |
- |
- |
PREDICTED: calcium-dependent protein kinase 8-like [Jatropha curcas] |
| 20 |
Hb_003883_010 |
0.1214774199 |
- |
- |
PREDICTED: cold-inducible RNA-binding protein [Jatropha curcas] |