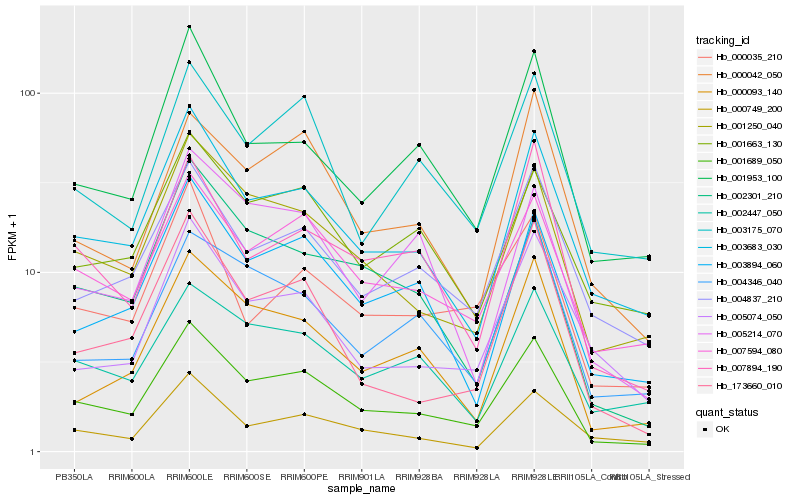
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001689_050 |
0.0 |
- |
- |
PREDICTED: cytochrome P450 CYP749A22-like [Prunus mume] |
| 2 |
Hb_001250_040 |
0.0878379314 |
- |
- |
hypothetical protein CISIN_1g018444mg [Citrus sinensis] |
| 3 |
Hb_003683_030 |
0.0889430331 |
- |
- |
PREDICTED: uridine-cytidine kinase C isoform X1 [Jatropha curcas] |
| 4 |
Hb_007594_080 |
0.0923040433 |
- |
- |
PREDICTED: polygalacturonate 4-alpha-galacturonosyltransferase [Jatropha curcas] |
| 5 |
Hb_003894_060 |
0.1141267874 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_004837_210 |
0.1168995847 |
- |
- |
PREDICTED: calcium-dependent protein kinase 8-like [Jatropha curcas] |
| 7 |
Hb_001953_100 |
0.1210970312 |
- |
- |
PREDICTED: glutamate-1-semialdehyde 2,1-aminomutase 2, chloroplastic [Jatropha curcas] |
| 8 |
Hb_003175_070 |
0.1217162745 |
- |
- |
pyrophosphate-dependent phosphofructokinase, partial [Hevea brasiliensis] |
| 9 |
Hb_002447_050 |
0.1221246806 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 10 |
Hb_001663_130 |
0.1243899522 |
- |
- |
acyl-CoA thioesterase, putative [Ricinus communis] |
| 11 |
Hb_007894_190 |
0.1249934158 |
- |
- |
PREDICTED: uncharacterized protein LOC105632935 [Jatropha curcas] |
| 12 |
Hb_002301_210 |
0.125082651 |
- |
- |
PREDICTED: receptor-like protein kinase At5g59670 isoform X2 [Jatropha curcas] |
| 13 |
Hb_173660_010 |
0.125676138 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Malus domestica] |
| 14 |
Hb_000093_140 |
0.1257116219 |
- |
- |
PREDICTED: fructokinase-like 2, chloroplastic [Jatropha curcas] |
| 15 |
Hb_000035_210 |
0.1291394506 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein NPY1 [Jatropha curcas] |
| 16 |
Hb_000042_050 |
0.1300059871 |
- |
- |
PREDICTED: uncharacterized protein LOC105632791 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000749_200 |
0.1317494039 |
- |
- |
PREDICTED: plant intracellular Ras-group-related LRR protein 6 isoform X1 [Jatropha curcas] |
| 18 |
Hb_005074_050 |
0.1334683614 |
- |
- |
PREDICTED: myosin-6-like [Jatropha curcas] |
| 19 |
Hb_004346_040 |
0.1344476918 |
- |
- |
hypothetical protein POPTR_0016s06630g [Populus trichocarpa] |
| 20 |
Hb_005214_070 |
0.1352414311 |
- |
- |
PREDICTED: fatty acid amide hydrolase [Jatropha curcas] |