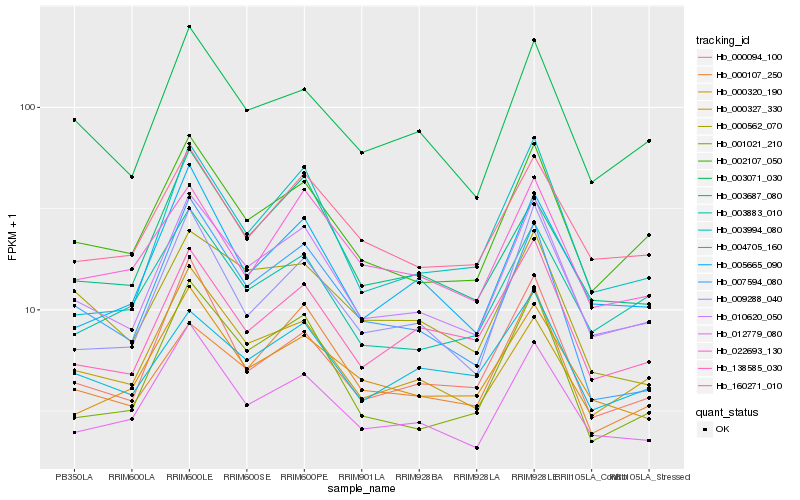
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010620_050 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105646119 [Jatropha curcas] |
| 2 |
Hb_002107_050 |
0.0624568192 |
- |
- |
PREDICTED: prostaglandin E synthase 2 [Jatropha curcas] |
| 3 |
Hb_012779_080 |
0.0677628112 |
- |
- |
PREDICTED: carbon catabolite repressor protein 4 homolog 3 isoform X3 [Jatropha curcas] |
| 4 |
Hb_003071_030 |
0.0706020186 |
- |
- |
PREDICTED: 20 kDa chaperonin, chloroplastic [Jatropha curcas] |
| 5 |
Hb_160271_010 |
0.0800967422 |
- |
- |
PREDICTED: FAD synthase-like isoform X2 [Jatropha curcas] |
| 6 |
Hb_138585_030 |
0.0802519517 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g54610 [Jatropha curcas] |
| 7 |
Hb_009288_040 |
0.0813169638 |
- |
- |
PREDICTED: uncharacterized protein LOC105648677 [Jatropha curcas] |
| 8 |
Hb_005665_090 |
0.0841803386 |
- |
- |
PREDICTED: 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase isoform X2 [Jatropha curcas] |
| 9 |
Hb_001021_210 |
0.0862529907 |
transcription factor |
TF Family: mTERF |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000320_190 |
0.0870704377 |
- |
- |
PREDICTED: aminomethyltransferase, mitochondrial [Jatropha curcas] |
| 11 |
Hb_003994_080 |
0.0876897629 |
- |
- |
PREDICTED: uncharacterized protein LOC105634384 [Jatropha curcas] |
| 12 |
Hb_007594_080 |
0.088430134 |
- |
- |
PREDICTED: polygalacturonate 4-alpha-galacturonosyltransferase [Jatropha curcas] |
| 13 |
Hb_003687_080 |
0.0884336917 |
- |
- |
PREDICTED: casein kinase I-like [Jatropha curcas] |
| 14 |
Hb_000094_100 |
0.0886983234 |
- |
- |
PREDICTED: probable methyltransferase PMT28 [Jatropha curcas] |
| 15 |
Hb_022693_130 |
0.0920142877 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 16 |
Hb_004705_160 |
0.0924531773 |
- |
- |
PREDICTED: protein MEI2-like 4 [Jatropha curcas] |
| 17 |
Hb_000107_250 |
0.0925684085 |
- |
- |
voltage-gated clc-type chloride channel, putative [Ricinus communis] |
| 18 |
Hb_000562_070 |
0.093315604 |
- |
- |
PREDICTED: uncharacterized protein LOC105635371 isoform X1 [Jatropha curcas] |
| 19 |
Hb_003883_010 |
0.0935668243 |
- |
- |
PREDICTED: cold-inducible RNA-binding protein [Jatropha curcas] |
| 20 |
Hb_000327_330 |
0.0943868596 |
- |
- |
conserved hypothetical protein [Ricinus communis] |