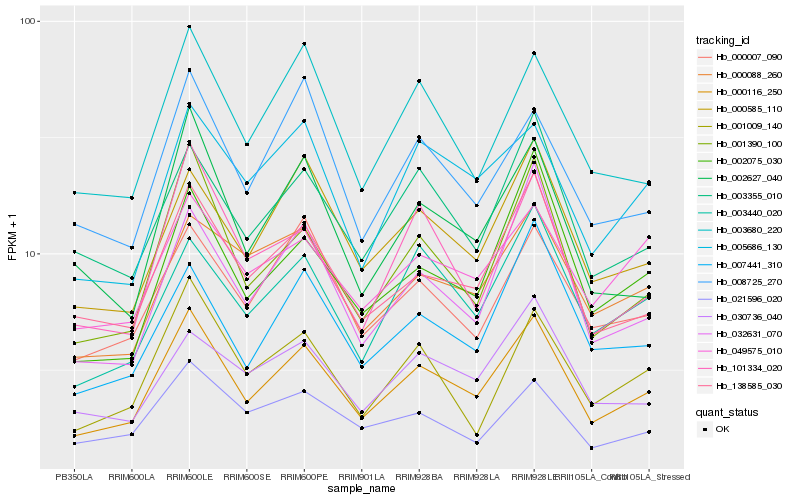
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000116_250 |
0.0 |
- |
- |
PREDICTED: DNA mismatch repair protein MLH3 [Jatropha curcas] |
| 2 |
Hb_138585_030 |
0.0761457922 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g54610 [Jatropha curcas] |
| 3 |
Hb_001390_100 |
0.0818941524 |
- |
- |
PREDICTED: LL-diaminopimelate aminotransferase, chloroplastic [Jatropha curcas] |
| 4 |
Hb_000585_110 |
0.0857285982 |
- |
- |
PREDICTED: probable protein phosphatase 2C 66 [Jatropha curcas] |
| 5 |
Hb_003355_010 |
0.0897888547 |
- |
- |
Heat shock 70 kDa protein, putative [Ricinus communis] |
| 6 |
Hb_003440_020 |
0.0899031869 |
- |
- |
PREDICTED: serine protease SPPA, chloroplastic isoform X2 [Jatropha curcas] |
| 7 |
Hb_000007_090 |
0.0906199018 |
- |
- |
PREDICTED: LAG1 longevity assurance homolog 3 [Jatropha curcas] |
| 8 |
Hb_003680_220 |
0.0931306622 |
- |
- |
hypothetical protein B456_002G243700 [Gossypium raimondii] |
| 9 |
Hb_005686_130 |
0.0941667687 |
rubber biosynthesis |
Gene Name: Isopentenyl-diphosphate delta isomerase |
isopentenyl pyrophosphate isomerase [Hevea brasiliensis] |
| 10 |
Hb_007441_310 |
0.094580537 |
- |
- |
PREDICTED: protein TRIGALACTOSYLDIACYLGLYCEROL 4, chloroplastic [Jatropha curcas] |
| 11 |
Hb_002075_030 |
0.0965876146 |
- |
- |
PREDICTED: glycerol-3-phosphate acyltransferase, chloroplastic [Jatropha curcas] |
| 12 |
Hb_000088_260 |
0.098066247 |
- |
- |
PREDICTED: uncharacterized protein LOC105636987 [Jatropha curcas] |
| 13 |
Hb_002627_040 |
0.0992313045 |
- |
- |
prolyl 4-hydroxylase alpha subunit, putative [Ricinus communis] |
| 14 |
Hb_001009_140 |
0.099267552 |
- |
- |
hypothetical protein JCGZ_11337 [Jatropha curcas] |
| 15 |
Hb_101334_020 |
0.0993978842 |
- |
- |
PREDICTED: uncharacterized WD repeat-containing protein C2A9.03-like [Jatropha curcas] |
| 16 |
Hb_049575_010 |
0.099641788 |
- |
- |
hypothetical protein POPTR_0001s15330g [Populus trichocarpa] |
| 17 |
Hb_030736_040 |
0.1006332746 |
- |
- |
PREDICTED: uncharacterized protein LOC105638926 isoform X1 [Jatropha curcas] |
| 18 |
Hb_021596_020 |
0.1006852273 |
- |
- |
hypothetical protein JCGZ_02034 [Jatropha curcas] |
| 19 |
Hb_032631_070 |
0.1009185147 |
- |
- |
PREDICTED: lisH domain and HEAT repeat-containing protein KIAA1468 homolog isoform X1 [Jatropha curcas] |
| 20 |
Hb_008725_270 |
0.1019741219 |
- |
- |
PREDICTED: probable protein disulfide-isomerase A6 [Jatropha curcas] |