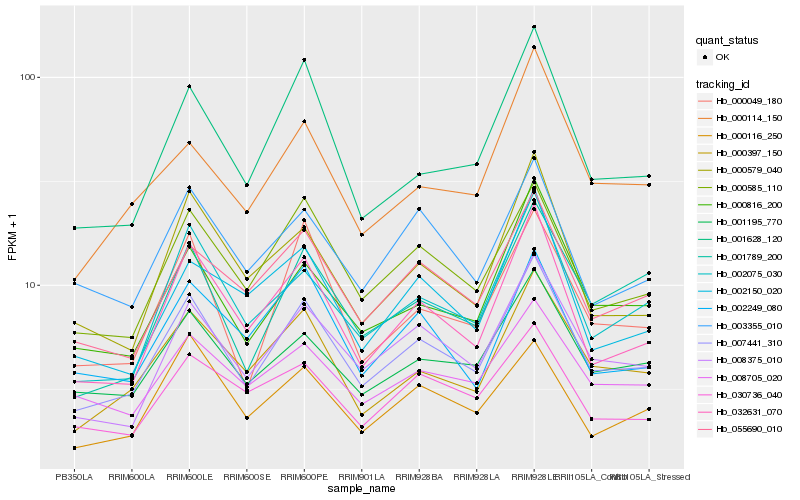
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007441_310 |
0.0 |
- |
- |
PREDICTED: protein TRIGALACTOSYLDIACYLGLYCEROL 4, chloroplastic [Jatropha curcas] |
| 2 |
Hb_008375_010 |
0.0637943783 |
- |
- |
PREDICTED: carbon catabolite repressor protein 4 homolog 4 isoform X3 [Pyrus x bretschneideri] |
| 3 |
Hb_032631_070 |
0.0681284984 |
- |
- |
PREDICTED: lisH domain and HEAT repeat-containing protein KIAA1468 homolog isoform X1 [Jatropha curcas] |
| 4 |
Hb_000585_110 |
0.069059941 |
- |
- |
PREDICTED: probable protein phosphatase 2C 66 [Jatropha curcas] |
| 5 |
Hb_000397_150 |
0.072428355 |
- |
- |
PREDICTED: probable flavin-containing monooxygenase 1 [Jatropha curcas] |
| 6 |
Hb_055690_010 |
0.0757610466 |
- |
- |
PREDICTED: pheophytinase, chloroplastic [Jatropha curcas] |
| 7 |
Hb_001195_770 |
0.076645506 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase ARI2 [Jatropha curcas] |
| 8 |
Hb_000816_200 |
0.0772194184 |
- |
- |
2-deoxyglucose-6-phosphate phosphatase, putative [Ricinus communis] |
| 9 |
Hb_002075_030 |
0.0793304052 |
- |
- |
PREDICTED: glycerol-3-phosphate acyltransferase, chloroplastic [Jatropha curcas] |
| 10 |
Hb_001628_120 |
0.0884140342 |
- |
- |
putative ascorbate peroxidase, partial [Taraxacum brevicorniculatum] |
| 11 |
Hb_000579_040 |
0.0907337036 |
- |
- |
PREDICTED: putative GTP-binding protein 6 [Jatropha curcas] |
| 12 |
Hb_000049_180 |
0.0908071258 |
- |
- |
aromatic amino acid decarboxylase, putative [Ricinus communis] |
| 13 |
Hb_008705_020 |
0.0911228608 |
- |
- |
PREDICTED: lisH domain and HEAT repeat-containing protein KIAA1468 homolog isoform X1 [Jatropha curcas] |
| 14 |
Hb_003355_010 |
0.0921848133 |
- |
- |
Heat shock 70 kDa protein, putative [Ricinus communis] |
| 15 |
Hb_001789_200 |
0.0943699282 |
- |
- |
PREDICTED: sec-independent protein translocase protein TATB, chloroplastic [Jatropha curcas] |
| 16 |
Hb_000116_250 |
0.094580537 |
- |
- |
PREDICTED: DNA mismatch repair protein MLH3 [Jatropha curcas] |
| 17 |
Hb_002249_080 |
0.0955817194 |
- |
- |
lysosomal alpha-mannosidase, putative [Ricinus communis] |
| 18 |
Hb_030736_040 |
0.0960192121 |
- |
- |
PREDICTED: uncharacterized protein LOC105638926 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000114_150 |
0.0962857715 |
rubber biosynthesis |
Gene Name: Geranyl geranyl diphosphate synthase |
geranylgeranyl-diphosphate synthase [Hevea brasiliensis] |
| 20 |
Hb_002150_020 |
0.0965618088 |
- |
- |
PREDICTED: translation initiation factor IF-2, mitochondrial isoform X1 [Jatropha curcas] |