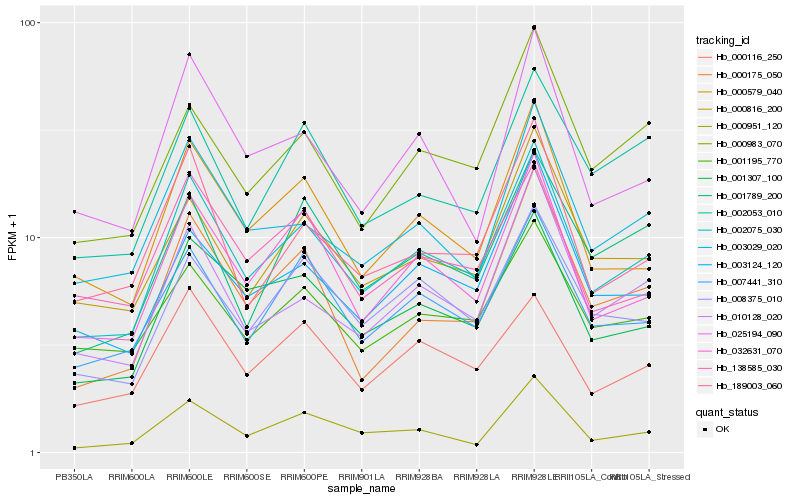
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002075_030 |
0.0 |
- |
- |
PREDICTED: glycerol-3-phosphate acyltransferase, chloroplastic [Jatropha curcas] |
| 2 |
Hb_032631_070 |
0.0741006873 |
- |
- |
PREDICTED: lisH domain and HEAT repeat-containing protein KIAA1468 homolog isoform X1 [Jatropha curcas] |
| 3 |
Hb_001307_100 |
0.0748341607 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |
| 4 |
Hb_000579_040 |
0.0766619948 |
- |
- |
PREDICTED: putative GTP-binding protein 6 [Jatropha curcas] |
| 5 |
Hb_007441_310 |
0.0793304052 |
- |
- |
PREDICTED: protein TRIGALACTOSYLDIACYLGLYCEROL 4, chloroplastic [Jatropha curcas] |
| 6 |
Hb_003029_020 |
0.0838043646 |
- |
- |
ATP-dependent Clp protease proteolytic subunit, putative [Ricinus communis] |
| 7 |
Hb_010128_020 |
0.0847424897 |
- |
- |
hypothetical protein POPTR_0001s24210g [Populus trichocarpa] |
| 8 |
Hb_000816_200 |
0.0876825758 |
- |
- |
2-deoxyglucose-6-phosphate phosphatase, putative [Ricinus communis] |
| 9 |
Hb_008375_010 |
0.0882103519 |
- |
- |
PREDICTED: carbon catabolite repressor protein 4 homolog 4 isoform X3 [Pyrus x bretschneideri] |
| 10 |
Hb_189003_060 |
0.0883170354 |
- |
- |
PREDICTED: uroporphyrinogen decarboxylase 1, chloroplastic isoform X1 [Jatropha curcas] |
| 11 |
Hb_001195_770 |
0.0903515141 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase ARI2 [Jatropha curcas] |
| 12 |
Hb_003124_120 |
0.0916843012 |
- |
- |
PREDICTED: fructose-1,6-bisphosphatase, cytosolic [Jatropha curcas] |
| 13 |
Hb_025194_090 |
0.0956157149 |
- |
- |
coproporphyrinogen III oxidase, putative [Ricinus communis] |
| 14 |
Hb_000116_250 |
0.0965876146 |
- |
- |
PREDICTED: DNA mismatch repair protein MLH3 [Jatropha curcas] |
| 15 |
Hb_000983_070 |
0.0969078512 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_138585_030 |
0.0986425947 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g54610 [Jatropha curcas] |
| 17 |
Hb_000175_050 |
0.1007361112 |
- |
- |
- |
| 18 |
Hb_002053_010 |
0.1040945314 |
- |
- |
PREDICTED: clp protease-related protein At4g12060, chloroplastic-like [Jatropha curcas] |
| 19 |
Hb_001789_200 |
0.105664804 |
- |
- |
PREDICTED: sec-independent protein translocase protein TATB, chloroplastic [Jatropha curcas] |
| 20 |
Hb_000951_120 |
0.1058602039 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105636019 [Jatropha curcas] |