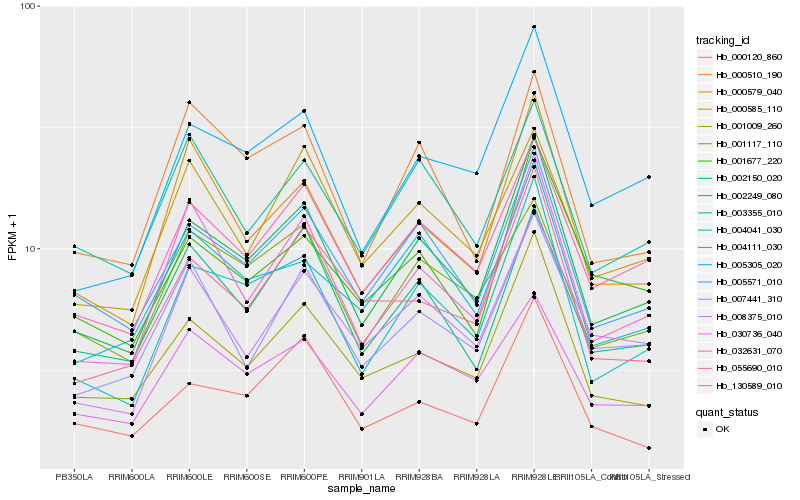
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002150_020 |
0.0 |
- |
- |
PREDICTED: translation initiation factor IF-2, mitochondrial isoform X1 [Jatropha curcas] |
| 2 |
Hb_004041_030 |
0.0480784771 |
- |
- |
PREDICTED: mitochondrial substrate carrier family protein C-like isoform X1 [Populus euphratica] |
| 3 |
Hb_005571_010 |
0.0634907189 |
- |
- |
PREDICTED: 15-cis-phytoene desaturase, chloroplastic/chromoplastic [Jatropha curcas] |
| 4 |
Hb_055690_010 |
0.0647612424 |
- |
- |
PREDICTED: pheophytinase, chloroplastic [Jatropha curcas] |
| 5 |
Hb_001009_260 |
0.0725602575 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase 1 isoform X1 [Jatropha curcas] |
| 6 |
Hb_032631_070 |
0.0748567827 |
- |
- |
PREDICTED: lisH domain and HEAT repeat-containing protein KIAA1468 homolog isoform X1 [Jatropha curcas] |
| 7 |
Hb_005305_020 |
0.0753346983 |
- |
- |
PREDICTED: dicarboxylate transporter 1, chloroplastic [Jatropha curcas] |
| 8 |
Hb_030736_040 |
0.0850902165 |
- |
- |
PREDICTED: uncharacterized protein LOC105638926 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000579_040 |
0.0865743369 |
- |
- |
PREDICTED: putative GTP-binding protein 6 [Jatropha curcas] |
| 10 |
Hb_004111_030 |
0.0873547503 |
- |
- |
PREDICTED: DNA gyrase subunit B, chloroplastic/mitochondrial-like [Jatropha curcas] |
| 11 |
Hb_002249_080 |
0.0907091596 |
- |
- |
lysosomal alpha-mannosidase, putative [Ricinus communis] |
| 12 |
Hb_130589_010 |
0.0926091526 |
- |
- |
PREDICTED: clustered mitochondria protein homolog [Jatropha curcas] |
| 13 |
Hb_000120_860 |
0.0931885149 |
- |
- |
nucellin, putative [Ricinus communis] |
| 14 |
Hb_000510_190 |
0.0945189052 |
- |
- |
glutathione reductase [Hevea brasiliensis] |
| 15 |
Hb_008375_010 |
0.094971887 |
- |
- |
PREDICTED: carbon catabolite repressor protein 4 homolog 4 isoform X3 [Pyrus x bretschneideri] |
| 16 |
Hb_001677_220 |
0.0951385983 |
- |
- |
PREDICTED: folylpolyglutamate synthase [Jatropha curcas] |
| 17 |
Hb_001117_110 |
0.0953114836 |
- |
- |
PREDICTED: dynamin-2A [Jatropha curcas] |
| 18 |
Hb_007441_310 |
0.0965618088 |
- |
- |
PREDICTED: protein TRIGALACTOSYLDIACYLGLYCEROL 4, chloroplastic [Jatropha curcas] |
| 19 |
Hb_000585_110 |
0.0966139618 |
- |
- |
PREDICTED: probable protein phosphatase 2C 66 [Jatropha curcas] |
| 20 |
Hb_003355_010 |
0.0979928409 |
- |
- |
Heat shock 70 kDa protein, putative [Ricinus communis] |