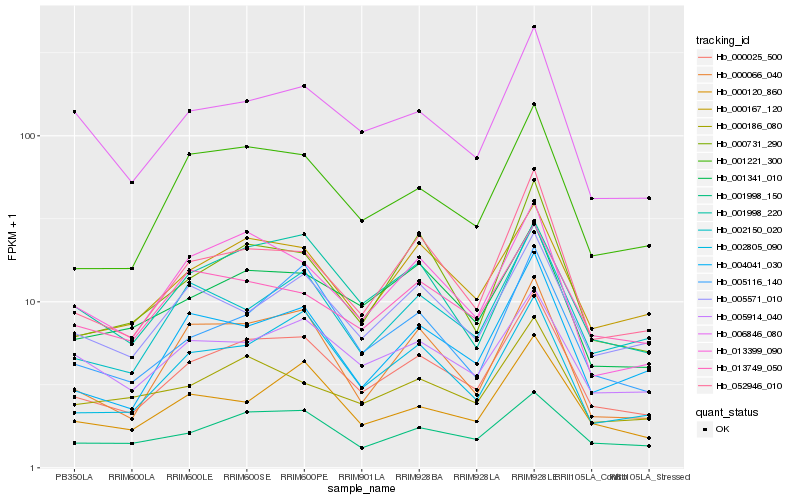
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000025_500 |
0.0 |
- |
- |
Ran GTPase binding protein, putative [Ricinus communis] |
| 2 |
Hb_000167_120 |
0.0829618197 |
- |
- |
PREDICTED: U-box domain-containing protein 14 [Jatropha curcas] |
| 3 |
Hb_001221_300 |
0.0836847093 |
- |
- |
PREDICTED: acetolactate synthase 3, chloroplastic [Jatropha curcas] |
| 4 |
Hb_052946_010 |
0.0848994351 |
- |
- |
PREDICTED: uncharacterized protein LOC105639103 [Jatropha curcas] |
| 5 |
Hb_001998_150 |
0.086970575 |
- |
- |
hypothetical protein JCGZ_09140 [Jatropha curcas] |
| 6 |
Hb_001341_010 |
0.0886339416 |
- |
- |
PREDICTED: carbamoyl-phosphate synthase large chain, chloroplastic [Jatropha curcas] |
| 7 |
Hb_004041_030 |
0.0923024284 |
- |
- |
PREDICTED: mitochondrial substrate carrier family protein C-like isoform X1 [Populus euphratica] |
| 8 |
Hb_013399_090 |
0.0940901565 |
- |
- |
PREDICTED: phospholipid:diacylglycerol acyltransferase 1-like [Jatropha curcas] |
| 9 |
Hb_006846_080 |
0.0946652801 |
- |
- |
calnexin, putative [Ricinus communis] |
| 10 |
Hb_002805_090 |
0.0966785361 |
- |
- |
Ran GTPase binding protein, putative [Ricinus communis] |
| 11 |
Hb_000731_290 |
0.0975392609 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 12 |
Hb_005914_040 |
0.0982132404 |
- |
- |
o-linked n-acetylglucosamine transferase, ogt, putative [Ricinus communis] |
| 13 |
Hb_001998_220 |
0.0985094524 |
- |
- |
PREDICTED: isoamylase 1, chloroplastic [Jatropha curcas] |
| 14 |
Hb_000120_860 |
0.099119763 |
- |
- |
nucellin, putative [Ricinus communis] |
| 15 |
Hb_002150_020 |
0.0997663117 |
- |
- |
PREDICTED: translation initiation factor IF-2, mitochondrial isoform X1 [Jatropha curcas] |
| 16 |
Hb_000066_040 |
0.1009891739 |
- |
- |
PREDICTED: CDPK-related kinase 7-like [Populus euphratica] |
| 17 |
Hb_000186_080 |
0.1016436763 |
- |
- |
PREDICTED: protein unc-45 homolog B-like [Gossypium raimondii] |
| 18 |
Hb_005571_010 |
0.1025086796 |
- |
- |
PREDICTED: 15-cis-phytoene desaturase, chloroplastic/chromoplastic [Jatropha curcas] |
| 19 |
Hb_005116_140 |
0.1026025857 |
- |
- |
PREDICTED: RAN GTPase-activating protein 1 [Jatropha curcas] |
| 20 |
Hb_013749_050 |
0.1029734928 |
- |
- |
PREDICTED: uncharacterized protein LOC105647765 isoform X2 [Jatropha curcas] |