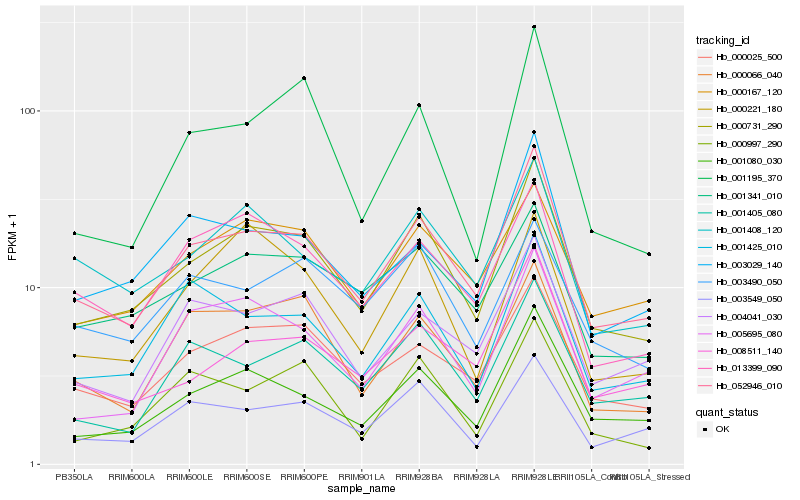
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000731_290 |
0.0 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 2 |
Hb_052946_010 |
0.0620900566 |
- |
- |
PREDICTED: uncharacterized protein LOC105639103 [Jatropha curcas] |
| 3 |
Hb_001080_030 |
0.0872405392 |
- |
- |
AMP dependent ligase, putative [Ricinus communis] |
| 4 |
Hb_000025_500 |
0.0975392609 |
- |
- |
Ran GTPase binding protein, putative [Ricinus communis] |
| 5 |
Hb_001341_010 |
0.1037646003 |
- |
- |
PREDICTED: carbamoyl-phosphate synthase large chain, chloroplastic [Jatropha curcas] |
| 6 |
Hb_000167_120 |
0.1067935765 |
- |
- |
PREDICTED: U-box domain-containing protein 14 [Jatropha curcas] |
| 7 |
Hb_003549_050 |
0.1102501797 |
- |
- |
PREDICTED: zinc phosphodiesterase ELAC protein 2-like [Jatropha curcas] |
| 8 |
Hb_001408_120 |
0.1118274203 |
- |
- |
dead box ATP-dependent RNA helicase, putative [Ricinus communis] |
| 9 |
Hb_005695_080 |
0.1163365745 |
- |
- |
PREDICTED: acetyl-CoA carboxylase 1-like [Jatropha curcas] |
| 10 |
Hb_013399_090 |
0.1173283428 |
- |
- |
PREDICTED: phospholipid:diacylglycerol acyltransferase 1-like [Jatropha curcas] |
| 11 |
Hb_000221_180 |
0.1222209472 |
- |
- |
VQ motif-containing family protein [Populus trichocarpa] |
| 12 |
Hb_000066_040 |
0.122551244 |
- |
- |
PREDICTED: CDPK-related kinase 7-like [Populus euphratica] |
| 13 |
Hb_001425_010 |
0.1227776023 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 14 |
Hb_000997_290 |
0.1227802196 |
- |
- |
PREDICTED: acylamino-acid-releasing enzyme [Jatropha curcas] |
| 15 |
Hb_001405_080 |
0.1230538809 |
- |
- |
PREDICTED: acylamino-acid-releasing enzyme [Jatropha curcas] |
| 16 |
Hb_008511_140 |
0.1247258888 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105632314 [Jatropha curcas] |
| 17 |
Hb_003029_140 |
0.1251389845 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g31400, chloroplastic [Jatropha curcas] |
| 18 |
Hb_001195_370 |
0.1257636692 |
- |
- |
- |
| 19 |
Hb_003490_050 |
0.1271985927 |
- |
- |
PREDICTED: probable cyclic nucleotide-gated ion channel 20, chloroplastic [Jatropha curcas] |
| 20 |
Hb_004041_030 |
0.1272559005 |
- |
- |
PREDICTED: mitochondrial substrate carrier family protein C-like isoform X1 [Populus euphratica] |