| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
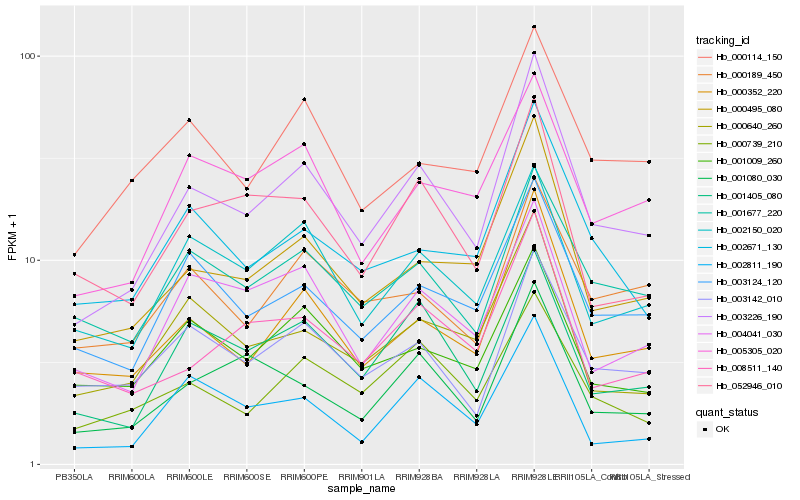
Hb_003226_190 |
0.0 |
- |
- |
PREDICTED: blue-light photoreceptor PHR2 [Jatropha curcas] |
| 2 |
Hb_000352_220 |
0.0876065087 |
- |
- |
PREDICTED: probable tyrosine--tRNA ligase, mitochondrial [Jatropha curcas] |
| 3 |
Hb_000495_080 |
0.095457359 |
- |
- |
Pentatricopeptide repeat (PPR-like) superfamily protein [Theobroma cacao] |
| 4 |
Hb_001405_080 |
0.1177295577 |
- |
- |
PREDICTED: acylamino-acid-releasing enzyme [Jatropha curcas] |
| 5 |
Hb_003124_120 |
0.1199115457 |
- |
- |
PREDICTED: fructose-1,6-bisphosphatase, cytosolic [Jatropha curcas] |
| 6 |
Hb_000640_260 |
0.1239660173 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g31400, chloroplastic [Jatropha curcas] |
| 7 |
Hb_001080_030 |
0.1244278233 |
- |
- |
AMP dependent ligase, putative [Ricinus communis] |
| 8 |
Hb_001677_220 |
0.1245438298 |
- |
- |
PREDICTED: folylpolyglutamate synthase [Jatropha curcas] |
| 9 |
Hb_002671_130 |
0.1252163257 |
- |
- |
GTP-binding protein typa/bipa, putative [Ricinus communis] |
| 10 |
Hb_003142_010 |
0.1259339866 |
transcription factor |
TF Family: SET |
PREDICTED: LOW QUALITY PROTEIN: histone-lysine N-methyltransferase ATX5-like [Populus euphratica] |
| 11 |
Hb_000739_210 |
0.1263487381 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g01970 [Jatropha curcas] |
| 12 |
Hb_005305_020 |
0.1277638398 |
- |
- |
PREDICTED: dicarboxylate transporter 1, chloroplastic [Jatropha curcas] |
| 13 |
Hb_008511_140 |
0.1280976993 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105632314 [Jatropha curcas] |
| 14 |
Hb_002150_020 |
0.1294706009 |
- |
- |
PREDICTED: translation initiation factor IF-2, mitochondrial isoform X1 [Jatropha curcas] |
| 15 |
Hb_052946_010 |
0.1313498721 |
- |
- |
PREDICTED: uncharacterized protein LOC105639103 [Jatropha curcas] |
| 16 |
Hb_004041_030 |
0.1316830605 |
- |
- |
PREDICTED: mitochondrial substrate carrier family protein C-like isoform X1 [Populus euphratica] |
| 17 |
Hb_000114_150 |
0.1350964736 |
rubber biosynthesis |
Gene Name: Geranyl geranyl diphosphate synthase |
geranylgeranyl-diphosphate synthase [Hevea brasiliensis] |
| 18 |
Hb_002811_190 |
0.135317162 |
- |
- |
tRNA pseudouridine synthase d, putative [Ricinus communis] |
| 19 |
Hb_000189_450 |
0.135540447 |
- |
- |
Zeta-carotene desaturase, chloroplast precursor, putative [Ricinus communis] |
| 20 |
Hb_001009_260 |
0.1361729176 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase 1 isoform X1 [Jatropha curcas] |