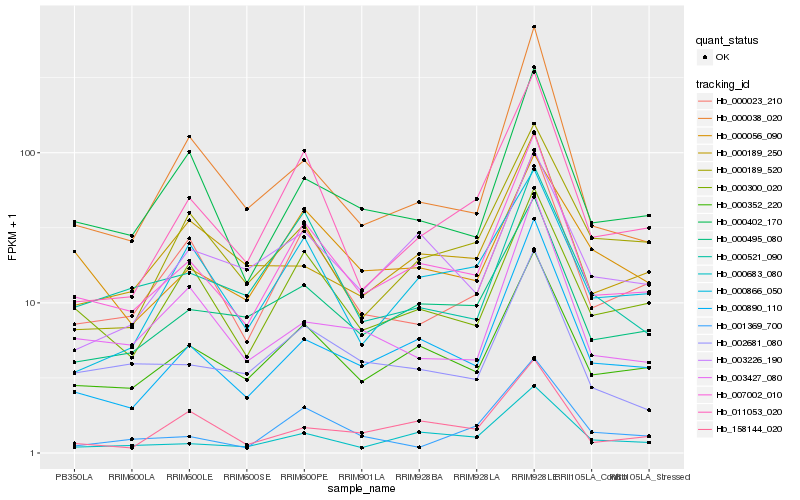
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007002_010 |
0.0 |
- |
- |
PREDICTED: dnaJ protein ERDJ3B [Jatropha curcas] |
| 2 |
Hb_000352_220 |
0.0821632581 |
- |
- |
PREDICTED: probable tyrosine--tRNA ligase, mitochondrial [Jatropha curcas] |
| 3 |
Hb_000890_110 |
0.0964018907 |
- |
- |
PREDICTED: GTP-binding protein OBGC, chloroplastic [Populus euphratica] |
| 4 |
Hb_000495_080 |
0.1074457673 |
- |
- |
Pentatricopeptide repeat (PPR-like) superfamily protein [Theobroma cacao] |
| 5 |
Hb_000683_080 |
0.1101430235 |
- |
- |
hypothetical protein EUGRSUZ_G013602, partial [Eucalyptus grandis] |
| 6 |
Hb_000023_210 |
0.1122180498 |
- |
- |
PREDICTED: protein TIC 55, chloroplastic [Jatropha curcas] |
| 7 |
Hb_011053_020 |
0.1155771459 |
- |
- |
lipoic acid synthetase, putative [Ricinus communis] |
| 8 |
Hb_000402_170 |
0.1208757143 |
- |
- |
PREDICTED: glutamate--glyoxylate aminotransferase 2 [Jatropha curcas] |
| 9 |
Hb_000300_020 |
0.1212170726 |
- |
- |
PREDICTED: grpE protein homolog, mitochondrial [Jatropha curcas] |
| 10 |
Hb_002681_080 |
0.1225852012 |
- |
- |
PREDICTED: uncharacterized protein LOC105633817 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000038_020 |
0.1424897418 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FTSH 2, chloroplastic [Jatropha curcas] |
| 12 |
Hb_003226_190 |
0.1454137579 |
- |
- |
PREDICTED: blue-light photoreceptor PHR2 [Jatropha curcas] |
| 13 |
Hb_000866_050 |
0.1461016661 |
- |
- |
phosphate transporter [Manihot esculenta] |
| 14 |
Hb_158144_020 |
0.146361292 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Vitis vinifera] |
| 15 |
Hb_003427_080 |
0.1464777793 |
- |
- |
PREDICTED: probable alanine--tRNA ligase, chloroplastic [Jatropha curcas] |
| 16 |
Hb_000189_250 |
0.14773695 |
- |
- |
PREDICTED: probable ribose-5-phosphate isomerase 3, chloroplastic [Jatropha curcas] |
| 17 |
Hb_000189_520 |
0.1490288096 |
- |
- |
PREDICTED: uncharacterized protein LOC105647658 [Jatropha curcas] |
| 18 |
Hb_000521_090 |
0.1509305219 |
- |
- |
PREDICTED: RING-H2 finger protein ATL5 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000056_090 |
0.1518477933 |
- |
- |
UDP-galactose transporter 3 isoform 1 [Theobroma cacao] |
| 20 |
Hb_001369_700 |
0.1519558054 |
- |
- |
- |