| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
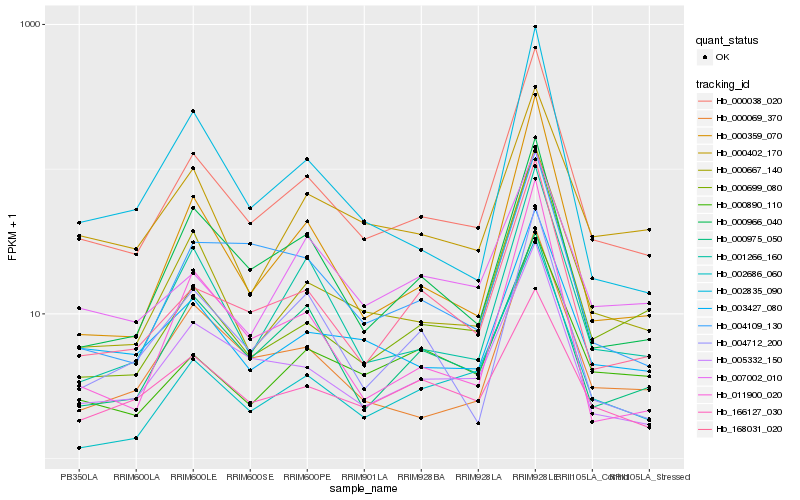
Hb_000038_020 |
0.0 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FTSH 2, chloroplastic [Jatropha curcas] |
| 2 |
Hb_000667_140 |
0.0837753482 |
- |
- |
PREDICTED: uridine kinase-like protein 3 isoform X1 [Jatropha curcas] |
| 3 |
Hb_168031_020 |
0.0922621383 |
- |
- |
PREDICTED: uncharacterized protein LOC105646684 isoform X2 [Jatropha curcas] |
| 4 |
Hb_000359_070 |
0.0986134809 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FTSH 2, chloroplastic [Jatropha curcas] |
| 5 |
Hb_005332_150 |
0.0990985708 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g46610 [Jatropha curcas] |
| 6 |
Hb_003427_080 |
0.1070857614 |
- |
- |
PREDICTED: probable alanine--tRNA ligase, chloroplastic [Jatropha curcas] |
| 7 |
Hb_000069_370 |
0.1142826906 |
- |
- |
- |
| 8 |
Hb_001266_160 |
0.1147555038 |
- |
- |
PREDICTED: pheophytinase, chloroplastic [Jatropha curcas] |
| 9 |
Hb_000966_040 |
0.1226326581 |
- |
- |
hypothetical protein JCGZ_01642 [Jatropha curcas] |
| 10 |
Hb_000890_110 |
0.1280092793 |
- |
- |
PREDICTED: GTP-binding protein OBGC, chloroplastic [Populus euphratica] |
| 11 |
Hb_002835_090 |
0.1298717677 |
- |
- |
RecName: Full=Elongation factor Tu, chloroplastic; Short=EF-Tu; Flags: Precursor [Glycine max] |
| 12 |
Hb_011900_020 |
0.1347505813 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g16390, chloroplastic [Jatropha curcas] |
| 13 |
Hb_000699_080 |
0.1360359171 |
- |
- |
PREDICTED: rubisco accumulation factor 1, chloroplastic [Jatropha curcas] |
| 14 |
Hb_002686_060 |
0.1364209418 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000402_170 |
0.137149975 |
- |
- |
PREDICTED: glutamate--glyoxylate aminotransferase 2 [Jatropha curcas] |
| 16 |
Hb_007002_010 |
0.1424897418 |
- |
- |
PREDICTED: dnaJ protein ERDJ3B [Jatropha curcas] |
| 17 |
Hb_004712_200 |
0.1454714938 |
- |
- |
hypothetical protein CICLE_v10010292mg [Citrus clementina] |
| 18 |
Hb_004109_130 |
0.1455178029 |
- |
- |
PREDICTED: translation initiation factor IF-2, chloroplastic [Jatropha curcas] |
| 19 |
Hb_000975_050 |
0.1462986454 |
- |
- |
PREDICTED: inner membrane protein PPF-1, chloroplastic [Jatropha curcas] |
| 20 |
Hb_166127_030 |
0.1463249341 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |