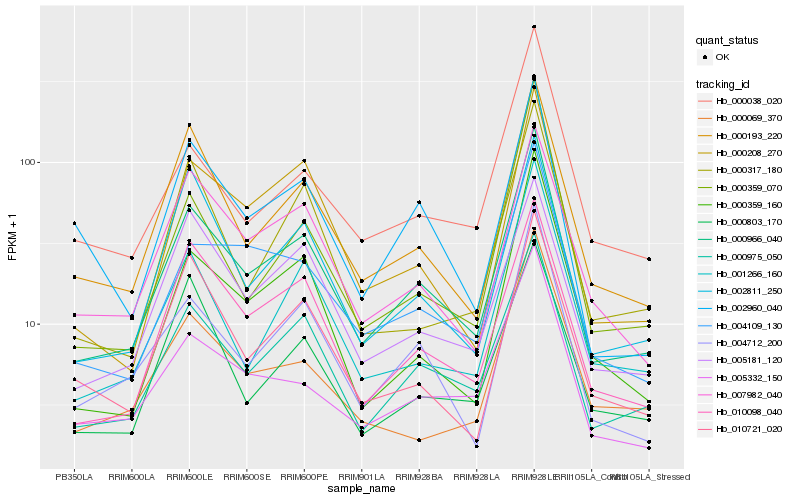
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000966_040 |
0.0 |
- |
- |
hypothetical protein JCGZ_01642 [Jatropha curcas] |
| 2 |
Hb_000975_050 |
0.0688616625 |
- |
- |
PREDICTED: inner membrane protein PPF-1, chloroplastic [Jatropha curcas] |
| 3 |
Hb_004712_200 |
0.0976909081 |
- |
- |
hypothetical protein CICLE_v10010292mg [Citrus clementina] |
| 4 |
Hb_000193_220 |
0.0979610755 |
- |
- |
PREDICTED: ribulose-phosphate 3-epimerase, chloroplastic [Jatropha curcas] |
| 5 |
Hb_010098_040 |
0.0991593179 |
- |
- |
PREDICTED: DNA-binding protein SMUBP-2 [Jatropha curcas] |
| 6 |
Hb_004109_130 |
0.1089358212 |
- |
- |
PREDICTED: translation initiation factor IF-2, chloroplastic [Jatropha curcas] |
| 7 |
Hb_000208_270 |
0.108973507 |
- |
- |
PREDICTED: malate dehydrogenase [NADP], chloroplastic [Jatropha curcas] |
| 8 |
Hb_005332_150 |
0.1096442664 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g46610 [Jatropha curcas] |
| 9 |
Hb_001266_160 |
0.1129761096 |
- |
- |
PREDICTED: pheophytinase, chloroplastic [Jatropha curcas] |
| 10 |
Hb_007982_040 |
0.1137105982 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase At1g79600, chloroplastic [Jatropha curcas] |
| 11 |
Hb_000359_160 |
0.113783856 |
- |
- |
PREDICTED: protein CAJ1 [Jatropha curcas] |
| 12 |
Hb_002811_250 |
0.1212485228 |
- |
- |
PREDICTED: probable ribose-5-phosphate isomerase 3, chloroplastic [Jatropha curcas] |
| 13 |
Hb_000038_020 |
0.1226326581 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FTSH 2, chloroplastic [Jatropha curcas] |
| 14 |
Hb_005181_120 |
0.1230929576 |
- |
- |
PREDICTED: psbB mRNA maturation factor Mbb1, chloroplastic isoform X1 [Jatropha curcas] |
| 15 |
Hb_002960_040 |
0.1238893433 |
- |
- |
PREDICTED: endoplasmin homolog [Jatropha curcas] |
| 16 |
Hb_010721_020 |
0.1245618544 |
- |
- |
PREDICTED: photosynthetic NDH subunit of lumenal location 4, chloroplastic [Jatropha curcas] |
| 17 |
Hb_000069_370 |
0.127950952 |
- |
- |
- |
| 18 |
Hb_000803_170 |
0.1301011078 |
- |
- |
PREDICTED: uncharacterized protein LOC105648312 [Jatropha curcas] |
| 19 |
Hb_000317_180 |
0.1310124842 |
- |
- |
PREDICTED: protein THYLAKOID FORMATION1, chloroplastic [Jatropha curcas] |
| 20 |
Hb_000359_070 |
0.1317974583 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FTSH 2, chloroplastic [Jatropha curcas] |