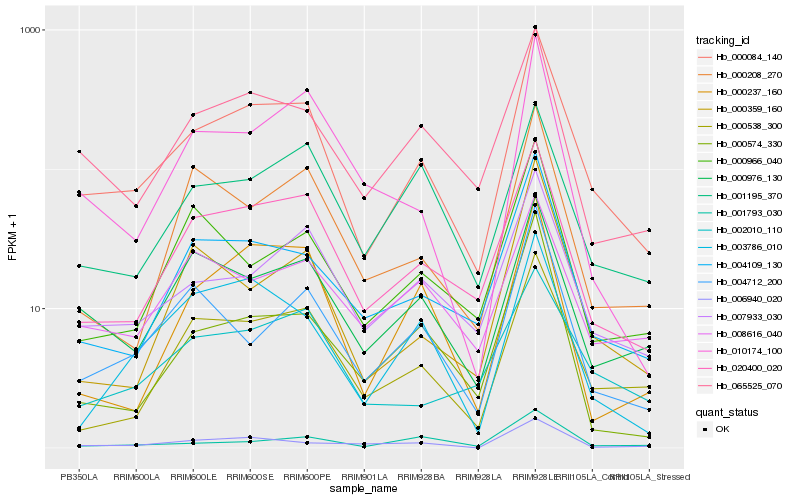
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000084_140 |
0.0 |
- |
- |
Aspartic proteinase nepenthesin-2 precursor, putative [Ricinus communis] |
| 2 |
Hb_020400_020 |
0.1177896381 |
- |
- |
starch synthase isoform II [Manihot esculenta] |
| 3 |
Hb_007933_030 |
0.1300819716 |
- |
- |
PREDICTED: uncharacterized protein LOC105639932 [Jatropha curcas] |
| 4 |
Hb_004109_130 |
0.1338994788 |
- |
- |
PREDICTED: translation initiation factor IF-2, chloroplastic [Jatropha curcas] |
| 5 |
Hb_065525_070 |
0.140400251 |
- |
- |
unnamed protein product [Homo sapiens] |
| 6 |
Hb_000538_300 |
0.1433425173 |
- |
- |
PREDICTED: WAT1-related protein At3g02690, chloroplastic [Jatropha curcas] |
| 7 |
Hb_000976_130 |
0.1498041329 |
- |
- |
PREDICTED: probable BOI-related E3 ubiquitin-protein ligase 2 [Jatropha curcas] |
| 8 |
Hb_002010_110 |
0.1606159441 |
- |
- |
PREDICTED: vacuolar amino acid transporter 1 [Jatropha curcas] |
| 9 |
Hb_004712_200 |
0.1617381645 |
- |
- |
hypothetical protein CICLE_v10010292mg [Citrus clementina] |
| 10 |
Hb_001793_030 |
0.1676036126 |
- |
- |
Cysteine-rich RLK (RECEPTOR-like protein kinase) 8 [Theobroma cacao] |
| 11 |
Hb_001195_370 |
0.1689366261 |
- |
- |
- |
| 12 |
Hb_008616_040 |
0.1699503904 |
- |
- |
PREDICTED: RNA-binding protein CP33, chloroplastic [Jatropha curcas] |
| 13 |
Hb_003786_010 |
0.1700024954 |
transcription factor |
TF Family: HB |
PREDICTED: BEL1-like homeodomain protein 2 isoform X2 [Jatropha curcas] |
| 14 |
Hb_000208_270 |
0.1700143992 |
- |
- |
PREDICTED: malate dehydrogenase [NADP], chloroplastic [Jatropha curcas] |
| 15 |
Hb_000359_160 |
0.1720459335 |
- |
- |
PREDICTED: protein CAJ1 [Jatropha curcas] |
| 16 |
Hb_010174_100 |
0.1723601751 |
- |
- |
PREDICTED: photosystem II core complex proteins psbY, chloroplastic-like [Populus euphratica] |
| 17 |
Hb_000966_040 |
0.1750387501 |
- |
- |
hypothetical protein JCGZ_01642 [Jatropha curcas] |
| 18 |
Hb_000574_330 |
0.1755437112 |
- |
- |
PREDICTED: clustered mitochondria protein homolog [Jatropha curcas] |
| 19 |
Hb_000237_160 |
0.1762894733 |
- |
- |
BEL1-like homeodomain protein 1 [Morus notabilis] |
| 20 |
Hb_006940_020 |
0.1767885185 |
- |
- |
polyprotein [Oryza australiensis] |