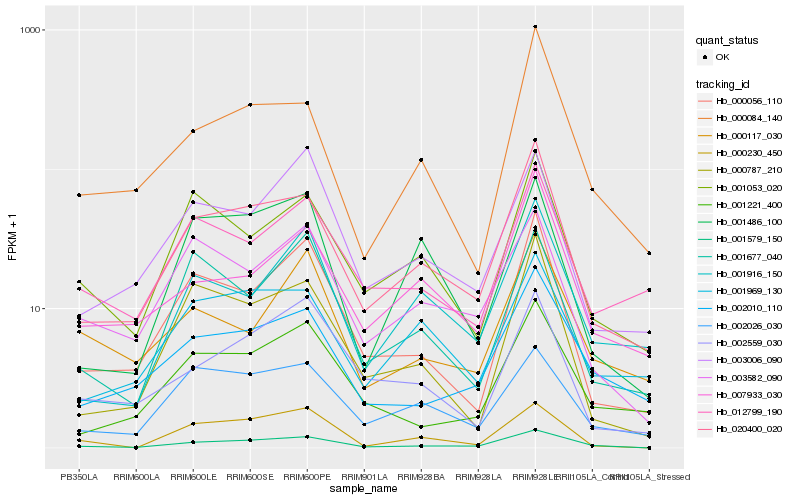
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001579_150 |
0.0 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 2 |
Hb_020400_020 |
0.1274857823 |
- |
- |
starch synthase isoform II [Manihot esculenta] |
| 3 |
Hb_002010_110 |
0.1458688755 |
- |
- |
PREDICTED: vacuolar amino acid transporter 1 [Jatropha curcas] |
| 4 |
Hb_000230_450 |
0.1599699853 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_003582_090 |
0.1649110314 |
- |
- |
PREDICTED: 65-kDa microtubule-associated protein 6-like [Jatropha curcas] |
| 6 |
Hb_002026_030 |
0.167868262 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: mitogen-activated protein kinase kinase kinase YODA [Jatropha curcas] |
| 7 |
Hb_001221_400 |
0.1685690591 |
- |
- |
PREDICTED: CBS domain-containing protein CBSCBSPB1 [Jatropha curcas] |
| 8 |
Hb_000056_110 |
0.169535536 |
- |
- |
PREDICTED: uncharacterized protein LOC105630405 [Jatropha curcas] |
| 9 |
Hb_000084_140 |
0.1780026902 |
- |
- |
Aspartic proteinase nepenthesin-2 precursor, putative [Ricinus communis] |
| 10 |
Hb_001053_020 |
0.1781470595 |
- |
- |
carotenoid cleavage dioxygenase 1 [Manihot esculenta] |
| 11 |
Hb_003006_090 |
0.1796951306 |
- |
- |
PREDICTED: cellulose synthase A catalytic subunit 2 [UDP-forming] isoform X2 [Jatropha curcas] |
| 12 |
Hb_000787_210 |
0.1846368971 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_002559_030 |
0.1854209651 |
- |
- |
oxidoreductase, putative [Ricinus communis] |
| 14 |
Hb_000117_030 |
0.1881491002 |
- |
- |
PREDICTED: protein ENHANCED DISEASE RESISTANCE 2 [Jatropha curcas] |
| 15 |
Hb_007933_030 |
0.188336712 |
- |
- |
PREDICTED: uncharacterized protein LOC105639932 [Jatropha curcas] |
| 16 |
Hb_001486_100 |
0.1884880108 |
- |
- |
tubulin alpha chain, putative [Ricinus communis] |
| 17 |
Hb_001916_150 |
0.1891344441 |
transcription factor |
TF Family: NF-YB |
PREDICTED: nuclear transcription factor Y subunit B-3 [Vitis vinifera] |
| 18 |
Hb_001677_040 |
0.1896662855 |
- |
- |
PREDICTED: ultraviolet-B receptor UVR8 isoform X2 [Jatropha curcas] |
| 19 |
Hb_001969_130 |
0.1898063999 |
- |
- |
PREDICTED: molybdate transporter 2 [Jatropha curcas] |
| 20 |
Hb_012799_190 |
0.190393106 |
- |
- |
PREDICTED: uncharacterized protein LOC105648490 [Jatropha curcas] |