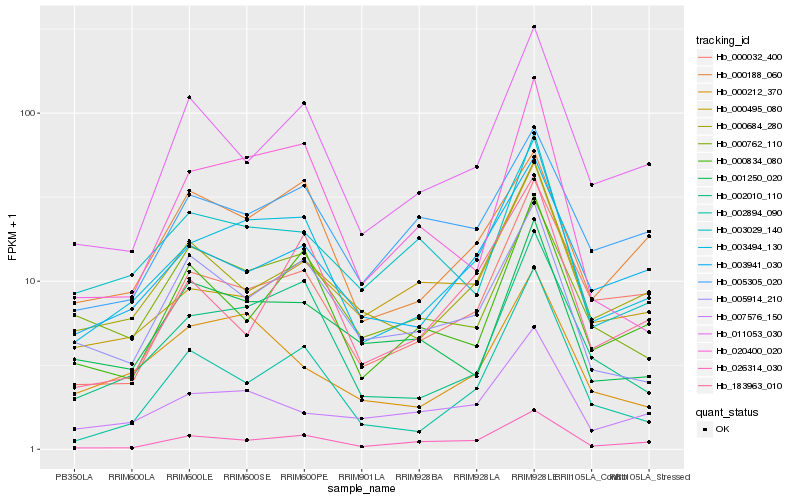
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003494_130 |
0.0 |
transcription factor |
TF Family: zf-HD |
PREDICTED: zinc-finger homeodomain protein 9-like [Jatropha curcas] |
| 2 |
Hb_002010_110 |
0.1114218117 |
- |
- |
PREDICTED: vacuolar amino acid transporter 1 [Jatropha curcas] |
| 3 |
Hb_003941_030 |
0.1336457895 |
- |
- |
Protease ecfE, putative [Ricinus communis] |
| 4 |
Hb_026314_030 |
0.1376699111 |
- |
- |
PREDICTED: transmembrane protein 41B [Jatropha curcas] |
| 5 |
Hb_000684_280 |
0.1426744154 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g09650, chloroplastic [Jatropha curcas] |
| 6 |
Hb_000032_400 |
0.1429659935 |
- |
- |
PREDICTED: UDP-glucose 4-epimerase GEPI48 [Jatropha curcas] |
| 7 |
Hb_011053_030 |
0.1447990143 |
- |
- |
- |
| 8 |
Hb_007576_150 |
0.1477624701 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g57430, chloroplastic [Jatropha curcas] |
| 9 |
Hb_020400_020 |
0.1496938172 |
- |
- |
starch synthase isoform II [Manihot esculenta] |
| 10 |
Hb_000212_370 |
0.1517988174 |
- |
- |
thioredoxin f-type, putative [Ricinus communis] |
| 11 |
Hb_000495_080 |
0.1559319763 |
- |
- |
Pentatricopeptide repeat (PPR-like) superfamily protein [Theobroma cacao] |
| 12 |
Hb_005914_210 |
0.155979301 |
- |
- |
PREDICTED: uncharacterized protein LOC105638411 [Jatropha curcas] |
| 13 |
Hb_000188_060 |
0.1576483424 |
- |
- |
PREDICTED: zinc finger A20 and AN1 domain-containing stress-associated protein 4-like [Jatropha curcas] |
| 14 |
Hb_002894_090 |
0.1580054181 |
- |
- |
PREDICTED: heparan-alpha-glucosaminide N-acetyltransferase [Jatropha curcas] |
| 15 |
Hb_000834_080 |
0.1606656581 |
- |
- |
PREDICTED: uncharacterized protein LOC105637718 [Jatropha curcas] |
| 16 |
Hb_003029_140 |
0.1610855879 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g31400, chloroplastic [Jatropha curcas] |
| 17 |
Hb_005305_020 |
0.1646575483 |
- |
- |
PREDICTED: dicarboxylate transporter 1, chloroplastic [Jatropha curcas] |
| 18 |
Hb_001250_020 |
0.1649426138 |
- |
- |
PREDICTED: CRS2-associated factor 2, chloroplastic [Jatropha curcas] |
| 19 |
Hb_000762_110 |
0.164955516 |
- |
- |
PREDICTED: protease Do-like 1, chloroplastic [Jatropha curcas] |
| 20 |
Hb_183963_010 |
0.1657081041 |
- |
- |
core region of GTP cyclohydrolase I family protein [Populus trichocarpa] |