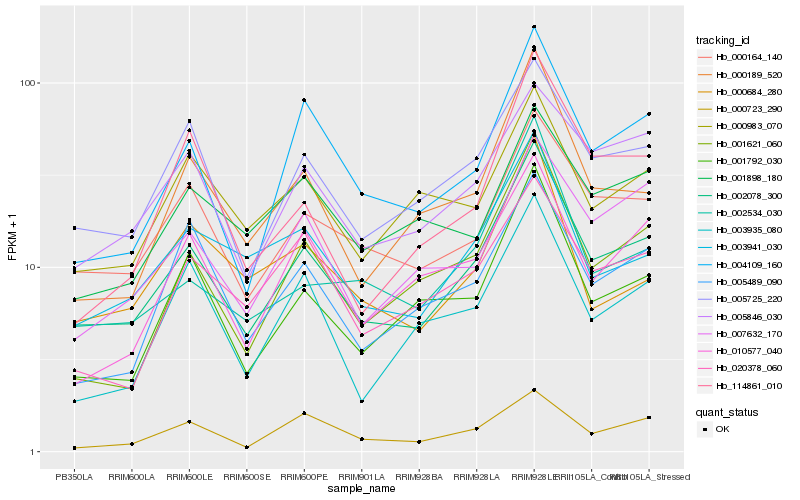
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002078_300 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105644817 isoform X1 [Jatropha curcas] |
| 2 |
Hb_005725_220 |
0.0922637159 |
- |
- |
PREDICTED: uncharacterized protein LOC105630808 [Jatropha curcas] |
| 3 |
Hb_000164_140 |
0.0933352818 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 4 |
Hb_004109_160 |
0.1045283054 |
- |
- |
PREDICTED: peptide methionine sulfoxide reductase B1, chloroplastic [Jatropha curcas] |
| 5 |
Hb_005846_030 |
0.1126574975 |
- |
- |
PREDICTED: 50S ribosomal protein L29, chloroplastic [Jatropha curcas] |
| 6 |
Hb_001792_030 |
0.1132936857 |
- |
- |
PREDICTED: aminoacyl tRNA synthase complex-interacting multifunctional protein 1 [Jatropha curcas] |
| 7 |
Hb_003941_030 |
0.1148558681 |
- |
- |
Protease ecfE, putative [Ricinus communis] |
| 8 |
Hb_000189_520 |
0.1151287108 |
- |
- |
PREDICTED: uncharacterized protein LOC105647658 [Jatropha curcas] |
| 9 |
Hb_001621_060 |
0.1161605569 |
- |
- |
PREDICTED: thylakoid lumenal 15 kDa protein 1, chloroplastic [Jatropha curcas] |
| 10 |
Hb_007632_170 |
0.1227730969 |
- |
- |
hypothetical protein JCGZ_01028 [Jatropha curcas] |
| 11 |
Hb_010577_040 |
0.1245726091 |
- |
- |
PREDICTED: uncharacterized protein LOC105643738 [Jatropha curcas] |
| 12 |
Hb_005489_090 |
0.1271156239 |
- |
- |
PREDICTED: peptidyl-tRNA hydrolase ICT1, mitochondrial [Jatropha curcas] |
| 13 |
Hb_000723_290 |
0.1293703851 |
- |
- |
calcium lipid binding protein, putative [Ricinus communis] |
| 14 |
Hb_003935_080 |
0.1303367531 |
- |
- |
PREDICTED: mechanosensitive ion channel protein 2, chloroplastic [Jatropha curcas] |
| 15 |
Hb_020378_060 |
0.1303876445 |
- |
- |
PREDICTED: methyltransferase-like protein 13 [Jatropha curcas] |
| 16 |
Hb_000983_070 |
0.1304926233 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_002534_030 |
0.1341570172 |
transcription factor |
TF Family: M-type |
mads box protein, putative [Ricinus communis] |
| 18 |
Hb_001898_180 |
0.1351408791 |
- |
- |
PREDICTED: translation initiation factor IF-1, chloroplastic [Jatropha curcas] |
| 19 |
Hb_000684_280 |
0.1356733692 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g09650, chloroplastic [Jatropha curcas] |
| 20 |
Hb_114861_010 |
0.1360933479 |
- |
- |
STRESS ENHANCED protein 1 [Populus trichocarpa] |