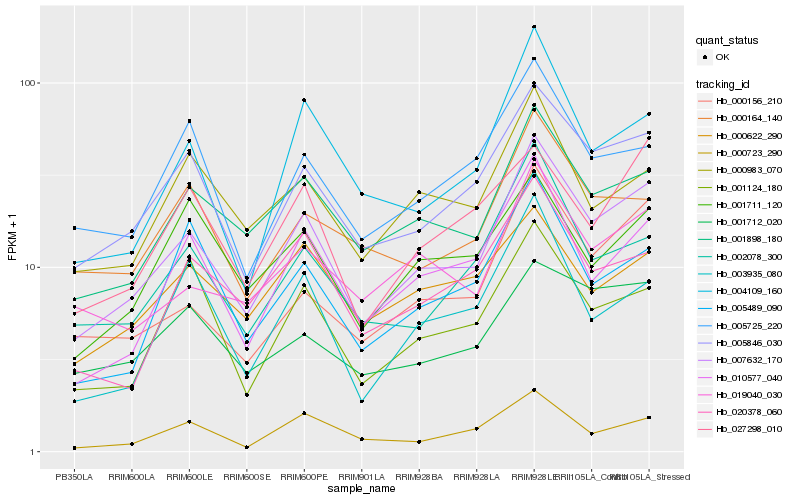
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007632_170 |
0.0 |
- |
- |
hypothetical protein JCGZ_01028 [Jatropha curcas] |
| 2 |
Hb_005846_030 |
0.0849600033 |
- |
- |
PREDICTED: 50S ribosomal protein L29, chloroplastic [Jatropha curcas] |
| 3 |
Hb_001898_180 |
0.0975538844 |
- |
- |
PREDICTED: translation initiation factor IF-1, chloroplastic [Jatropha curcas] |
| 4 |
Hb_010577_040 |
0.1085970331 |
- |
- |
PREDICTED: uncharacterized protein LOC105643738 [Jatropha curcas] |
| 5 |
Hb_019040_030 |
0.1185090731 |
- |
- |
PREDICTED: probable ribosome-binding factor A, chloroplastic [Jatropha curcas] |
| 6 |
Hb_000156_210 |
0.1193303616 |
- |
- |
PREDICTED: metal transporter Nramp1 [Jatropha curcas] |
| 7 |
Hb_002078_300 |
0.1227730969 |
- |
- |
PREDICTED: uncharacterized protein LOC105644817 isoform X1 [Jatropha curcas] |
| 8 |
Hb_004109_160 |
0.1227762193 |
- |
- |
PREDICTED: peptide methionine sulfoxide reductase B1, chloroplastic [Jatropha curcas] |
| 9 |
Hb_020378_060 |
0.1298856114 |
- |
- |
PREDICTED: methyltransferase-like protein 13 [Jatropha curcas] |
| 10 |
Hb_000723_290 |
0.1300328266 |
- |
- |
calcium lipid binding protein, putative [Ricinus communis] |
| 11 |
Hb_000983_070 |
0.1306511473 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_005725_220 |
0.1323958728 |
- |
- |
PREDICTED: uncharacterized protein LOC105630808 [Jatropha curcas] |
| 13 |
Hb_001124_180 |
0.1332593457 |
- |
- |
PREDICTED: uncharacterized protein LOC105171432 [Sesamum indicum] |
| 14 |
Hb_000164_140 |
0.1347816404 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 15 |
Hb_001712_020 |
0.1351279468 |
- |
- |
PREDICTED: uncharacterized protein LOC105641948 [Jatropha curcas] |
| 16 |
Hb_003935_080 |
0.1375350995 |
- |
- |
PREDICTED: mechanosensitive ion channel protein 2, chloroplastic [Jatropha curcas] |
| 17 |
Hb_027298_010 |
0.1378851332 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_005489_090 |
0.1387065442 |
- |
- |
PREDICTED: peptidyl-tRNA hydrolase ICT1, mitochondrial [Jatropha curcas] |
| 19 |
Hb_000622_290 |
0.1387900638 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At3g61590 [Jatropha curcas] |
| 20 |
Hb_001711_120 |
0.1403650713 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit 4, chloroplastic [Jatropha curcas] |