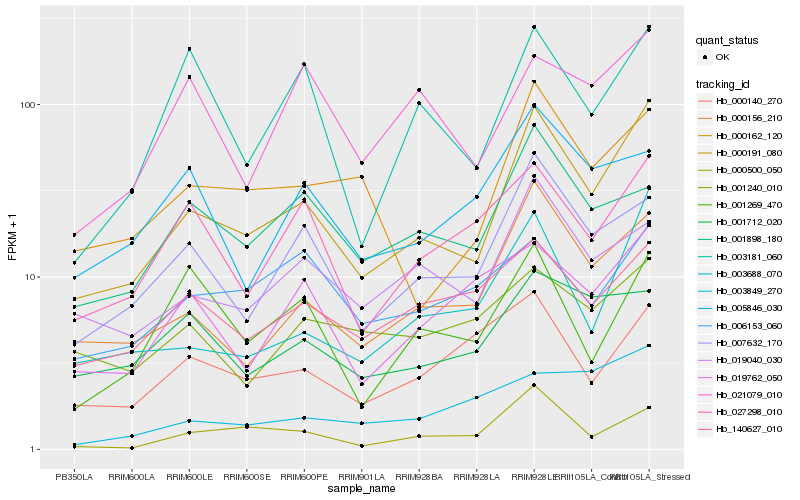
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000191_080 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105637945 [Jatropha curcas] |
| 2 |
Hb_000156_210 |
0.1329425319 |
- |
- |
PREDICTED: metal transporter Nramp1 [Jatropha curcas] |
| 3 |
Hb_003688_070 |
0.1468280977 |
- |
- |
PREDICTED: mitochondrial substrate carrier family protein E [Jatropha curcas] |
| 4 |
Hb_007632_170 |
0.1540589352 |
- |
- |
hypothetical protein JCGZ_01028 [Jatropha curcas] |
| 5 |
Hb_019040_030 |
0.169488826 |
- |
- |
PREDICTED: probable ribosome-binding factor A, chloroplastic [Jatropha curcas] |
| 6 |
Hb_000162_120 |
0.1703415085 |
- |
- |
PREDICTED: chaperone protein dnaJ 11, chloroplastic-like [Jatropha curcas] |
| 7 |
Hb_000500_050 |
0.1724063172 |
- |
- |
PREDICTED: uncharacterized protein LOC105160092 [Sesamum indicum] |
| 8 |
Hb_027298_010 |
0.1774176559 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_140627_010 |
0.1799260306 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_019762_050 |
0.1830126313 |
transcription factor |
TF Family: CSD |
PREDICTED: glycine-rich protein 2-like [Jatropha curcas] |
| 11 |
Hb_000140_270 |
0.1885358842 |
- |
- |
S-adenosyl-L-methionine-dependent methyltransferases superfamily protein [Theobroma cacao] |
| 12 |
Hb_003181_060 |
0.1888720714 |
- |
- |
PREDICTED: triosephosphate isomerase, cytosolic [Jatropha curcas] |
| 13 |
Hb_001712_020 |
0.1951500283 |
- |
- |
PREDICTED: uncharacterized protein LOC105641948 [Jatropha curcas] |
| 14 |
Hb_001898_180 |
0.195195203 |
- |
- |
PREDICTED: translation initiation factor IF-1, chloroplastic [Jatropha curcas] |
| 15 |
Hb_001269_470 |
0.1967036297 |
- |
- |
PREDICTED: uncharacterized protein LOC105630313 [Jatropha curcas] |
| 16 |
Hb_001240_010 |
0.196959087 |
- |
- |
PREDICTED: uncharacterized protein LOC105634101 [Jatropha curcas] |
| 17 |
Hb_006153_060 |
0.1999598256 |
- |
- |
catalytic, putative [Ricinus communis] |
| 18 |
Hb_021079_010 |
0.2014151293 |
- |
- |
PREDICTED: histone H2B-like [Populus euphratica] |
| 19 |
Hb_005846_030 |
0.2014680318 |
- |
- |
PREDICTED: 50S ribosomal protein L29, chloroplastic [Jatropha curcas] |
| 20 |
Hb_003849_270 |
0.2050853703 |
- |
- |
- |