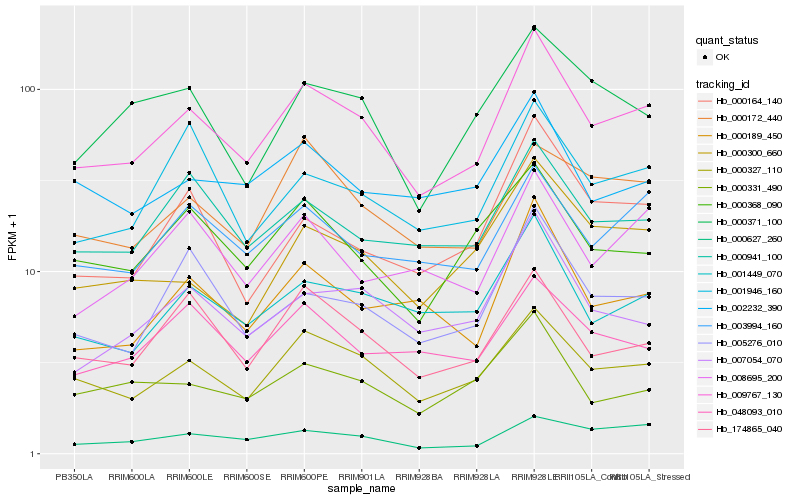
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009767_130 |
0.0 |
- |
- |
PREDICTED: preprotein translocase subunit SECE1 [Jatropha curcas] |
| 2 |
Hb_000327_110 |
0.0773767771 |
- |
- |
hypothetical protein JCGZ_06657 [Jatropha curcas] |
| 3 |
Hb_001449_070 |
0.0989170031 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000189_450 |
0.1076572938 |
- |
- |
Zeta-carotene desaturase, chloroplast precursor, putative [Ricinus communis] |
| 5 |
Hb_003994_160 |
0.1105321776 |
- |
- |
PREDICTED: uncharacterized protein LOC105131691 isoform X5 [Populus euphratica] |
| 6 |
Hb_000941_100 |
0.1109497279 |
- |
- |
thioredoxin-like 5 mRNA family protein [Populus trichocarpa] |
| 7 |
Hb_007054_070 |
0.1115097927 |
- |
- |
PREDICTED: probable monodehydroascorbate reductase, cytoplasmic isoform 2 [Jatropha curcas] |
| 8 |
Hb_174865_040 |
0.111514095 |
- |
- |
PREDICTED: crt homolog 1 isoform X2 [Jatropha curcas] |
| 9 |
Hb_005276_010 |
0.113451922 |
- |
- |
hypothetical protein CICLE_v10021605mg [Citrus clementina] |
| 10 |
Hb_000627_260 |
0.113775226 |
- |
- |
chloride channel clc, putative [Ricinus communis] |
| 11 |
Hb_000368_090 |
0.1176918462 |
- |
- |
TPA: hypothetical protein ZEAMMB73_442906 [Zea mays] |
| 12 |
Hb_000300_660 |
0.1188132833 |
- |
- |
unknown [Glycine max] |
| 13 |
Hb_000331_490 |
0.1194197305 |
- |
- |
PREDICTED: radical S-adenosyl methionine domain-containing protein 1, mitochondrial [Jatropha curcas] |
| 14 |
Hb_008695_200 |
0.1194276187 |
- |
- |
Protein grpE, putative [Ricinus communis] |
| 15 |
Hb_001946_160 |
0.1205238055 |
- |
- |
putative chaperon P13.9 [Castanea sativa] |
| 16 |
Hb_002232_390 |
0.121472622 |
- |
- |
PREDICTED: uncharacterized protein LOC105635994 [Jatropha curcas] |
| 17 |
Hb_048093_010 |
0.1225106124 |
- |
- |
PREDICTED: calcium uniporter protein 6, mitochondrial-like [Jatropha curcas] |
| 18 |
Hb_000371_100 |
0.1227804419 |
rubber biosynthesis |
Gene Name: Geranyl geranyl diphosphate synthase |
RecName: Full=Geranylgeranyl pyrophosphate synthase, chloroplastic; Short=GGPP synthase; Short=GGPS; AltName: Full=(2E,6E)-farnesyl diphosphate synthase; AltName: Full=Dimethylallyltranstransferase; AltName: Full=Farnesyl diphosphate synthase; AltName: Full=Farnesyltranstransferase; AltName: Full=Geranyltranstransferase; Flags: Precursor [Hevea brasiliensis] |
| 19 |
Hb_000172_440 |
0.1229934014 |
- |
- |
PREDICTED: NADPH-dependent 1-acyldihydroxyacetone phosphate reductase [Jatropha curcas] |
| 20 |
Hb_000164_140 |
0.1238767526 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |