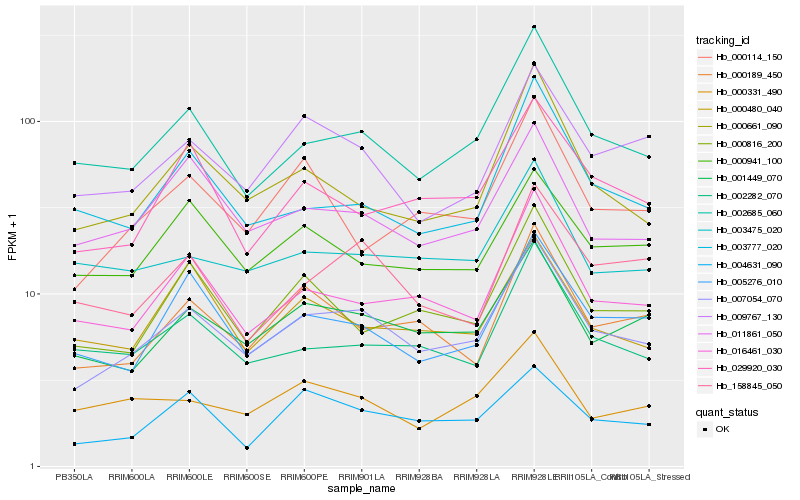
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007054_070 |
0.0 |
- |
- |
PREDICTED: probable monodehydroascorbate reductase, cytoplasmic isoform 2 [Jatropha curcas] |
| 2 |
Hb_001449_070 |
0.0911632941 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_002685_060 |
0.0923876405 |
- |
- |
30S ribosomal protein S5, putative [Ricinus communis] |
| 4 |
Hb_016461_030 |
0.1030052748 |
- |
- |
PREDICTED: putative leucine--tRNA ligase, mitochondrial isoform X1 [Jatropha curcas] |
| 5 |
Hb_000189_450 |
0.1082164851 |
- |
- |
Zeta-carotene desaturase, chloroplast precursor, putative [Ricinus communis] |
| 6 |
Hb_005276_010 |
0.1115050966 |
- |
- |
hypothetical protein CICLE_v10021605mg [Citrus clementina] |
| 7 |
Hb_009767_130 |
0.1115097927 |
- |
- |
PREDICTED: preprotein translocase subunit SECE1 [Jatropha curcas] |
| 8 |
Hb_000661_090 |
0.1116886564 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase At1g79600, chloroplastic [Jatropha curcas] |
| 9 |
Hb_011861_050 |
0.1120179068 |
- |
- |
PREDICTED: COBW domain-containing protein 1 [Jatropha curcas] |
| 10 |
Hb_002282_070 |
0.11375773 |
- |
- |
PREDICTED: protein DJ-1 homolog C isoform X2 [Jatropha curcas] |
| 11 |
Hb_029920_030 |
0.1150426784 |
- |
- |
PREDICTED: 2-Cys peroxiredoxin BAS1, chloroplastic-like [Jatropha curcas] |
| 12 |
Hb_000816_200 |
0.1164749603 |
- |
- |
2-deoxyglucose-6-phosphate phosphatase, putative [Ricinus communis] |
| 13 |
Hb_003475_020 |
0.1184536188 |
- |
- |
hypothetical protein JCGZ_13177 [Jatropha curcas] |
| 14 |
Hb_004631_090 |
0.1203997396 |
- |
- |
PREDICTED: probable glucan 1,3-beta-glucosidase A [Jatropha curcas] |
| 15 |
Hb_000941_100 |
0.1220765512 |
- |
- |
thioredoxin-like 5 mRNA family protein [Populus trichocarpa] |
| 16 |
Hb_158845_050 |
0.1241539556 |
- |
- |
PREDICTED: uncharacterized protein LOC105122890 [Populus euphratica] |
| 17 |
Hb_003777_020 |
0.1244207805 |
- |
- |
Fructose-1,6-bisphosphatase, cytosolic, putative [Ricinus communis] |
| 18 |
Hb_000114_150 |
0.12519665 |
rubber biosynthesis |
Gene Name: Geranyl geranyl diphosphate synthase |
geranylgeranyl-diphosphate synthase [Hevea brasiliensis] |
| 19 |
Hb_000480_040 |
0.1258532264 |
- |
- |
PREDICTED: L-Ala-D/L-amino acid epimerase isoform X2 [Jatropha curcas] |
| 20 |
Hb_000331_490 |
0.1262817217 |
- |
- |
PREDICTED: radical S-adenosyl methionine domain-containing protein 1, mitochondrial [Jatropha curcas] |