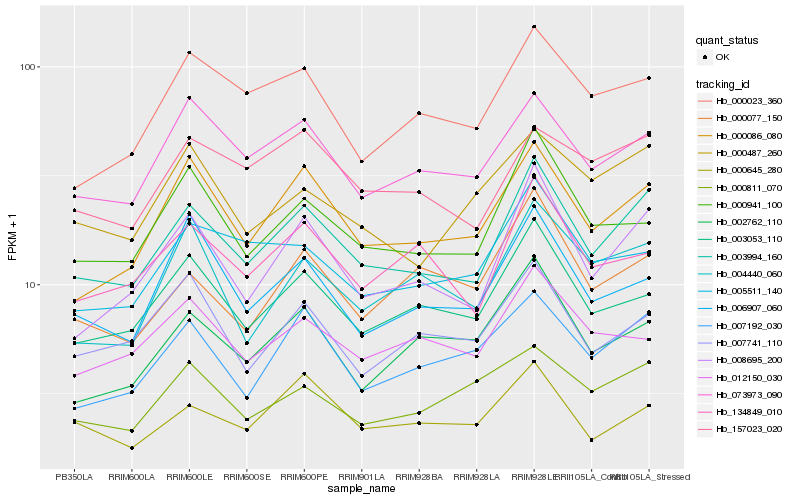
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003994_160 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105131691 isoform X5 [Populus euphratica] |
| 2 |
Hb_008695_200 |
0.0678701207 |
- |
- |
Protein grpE, putative [Ricinus communis] |
| 3 |
Hb_000086_080 |
0.0821768327 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_073973_090 |
0.0880788831 |
- |
- |
Aminotransferase ybdL, putative [Ricinus communis] |
| 5 |
Hb_003053_110 |
0.0887471895 |
- |
- |
PREDICTED: protease Do-like 1, chloroplastic [Jatropha curcas] |
| 6 |
Hb_007192_030 |
0.093382672 |
- |
- |
PREDICTED: manganese-dependent ADP-ribose/CDP-alcohol diphosphatase [Jatropha curcas] |
| 7 |
Hb_000941_100 |
0.0951163007 |
- |
- |
thioredoxin-like 5 mRNA family protein [Populus trichocarpa] |
| 8 |
Hb_157023_020 |
0.0963705395 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 9 |
Hb_000023_360 |
0.0997128942 |
- |
- |
phospholipid hydroperoxide glutathione peroxidase 1, chloroplastic [Jatropha curcas] |
| 10 |
Hb_134849_010 |
0.1003065246 |
- |
- |
ATP synthase subunit d, putative [Ricinus communis] |
| 11 |
Hb_000645_280 |
0.1021164299 |
- |
- |
PREDICTED: AMSH-like ubiquitin thioesterase 3 [Jatropha curcas] |
| 12 |
Hb_000487_260 |
0.1033538201 |
- |
- |
PREDICTED: protein Iojap, chloroplastic [Jatropha curcas] |
| 13 |
Hb_005511_140 |
0.1036352708 |
- |
- |
Ubiquinone biosynthesis protein coq-8, putative [Ricinus communis] |
| 14 |
Hb_002762_110 |
0.1053508404 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000811_070 |
0.1053988943 |
- |
- |
PREDICTED: uncharacterized protein LOC105650695 [Jatropha curcas] |
| 16 |
Hb_004440_060 |
0.1064376239 |
- |
- |
aldose 1-epimerase, putative [Ricinus communis] |
| 17 |
Hb_006907_060 |
0.1067954465 |
- |
- |
Alternative oxidase 4, chloroplast precursor, putative [Ricinus communis] |
| 18 |
Hb_000077_150 |
0.1073018235 |
- |
- |
PREDICTED: riboflavin synthase [Jatropha curcas] |
| 19 |
Hb_007741_110 |
0.1079128806 |
- |
- |
- |
| 20 |
Hb_012150_030 |
0.1084587765 |
- |
- |
PREDICTED: aspartate--tRNA ligase, mitochondrial [Jatropha curcas] |