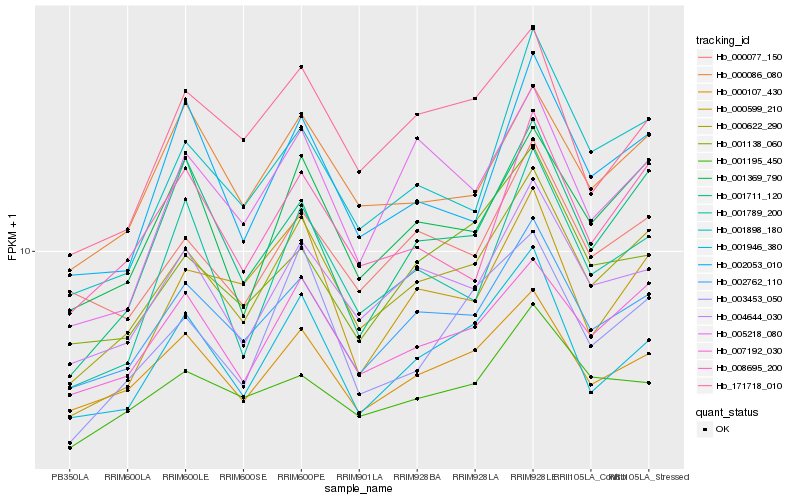
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000622_290 |
0.0 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At3g61590 [Jatropha curcas] |
| 2 |
Hb_002762_110 |
0.0581437363 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_007192_030 |
0.0710673809 |
- |
- |
PREDICTED: manganese-dependent ADP-ribose/CDP-alcohol diphosphatase [Jatropha curcas] |
| 4 |
Hb_004644_030 |
0.0714811152 |
- |
- |
PREDICTED: FAD synthase isoform X3 [Jatropha curcas] |
| 5 |
Hb_001946_380 |
0.0835136553 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 34-like [Jatropha curcas] |
| 6 |
Hb_000107_430 |
0.0838513522 |
- |
- |
PREDICTED: brefeldin A-inhibited guanine nucleotide-exchange protein 1 [Jatropha curcas] |
| 7 |
Hb_001369_790 |
0.0840802294 |
- |
- |
PREDICTED: clp protease-related protein At4g12060, chloroplastic [Jatropha curcas] |
| 8 |
Hb_000077_150 |
0.0882896427 |
- |
- |
PREDICTED: riboflavin synthase [Jatropha curcas] |
| 9 |
Hb_000086_080 |
0.096953031 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_003453_050 |
0.0969646445 |
- |
- |
PREDICTED: calcium-binding mitochondrial carrier protein SCaMC-3-like [Jatropha curcas] |
| 11 |
Hb_008695_200 |
0.0973087842 |
- |
- |
Protein grpE, putative [Ricinus communis] |
| 12 |
Hb_171718_010 |
0.0975166261 |
- |
- |
PREDICTED: disease resistance protein At4g27190-like [Eucalyptus grandis] |
| 13 |
Hb_001898_180 |
0.0981138537 |
- |
- |
PREDICTED: translation initiation factor IF-1, chloroplastic [Jatropha curcas] |
| 14 |
Hb_001711_120 |
0.0988676571 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit 4, chloroplastic [Jatropha curcas] |
| 15 |
Hb_005218_080 |
0.0991215754 |
- |
- |
PREDICTED: branched-chain-amino-acid aminotransferase 3, chloroplastic isoform X1 [Jatropha curcas] |
| 16 |
Hb_001195_450 |
0.1000881237 |
- |
- |
PREDICTED: uncharacterized protein LOC105633771 [Jatropha curcas] |
| 17 |
Hb_001138_060 |
0.1028464756 |
- |
- |
hypothetical protein CICLE_v10028086mg [Citrus clementina] |
| 18 |
Hb_001789_200 |
0.1034821394 |
- |
- |
PREDICTED: sec-independent protein translocase protein TATB, chloroplastic [Jatropha curcas] |
| 19 |
Hb_002053_010 |
0.1039329704 |
- |
- |
PREDICTED: clp protease-related protein At4g12060, chloroplastic-like [Jatropha curcas] |
| 20 |
Hb_000599_210 |
0.1039794204 |
- |
- |
serine/threonine-protein kinase, putative [Ricinus communis] |