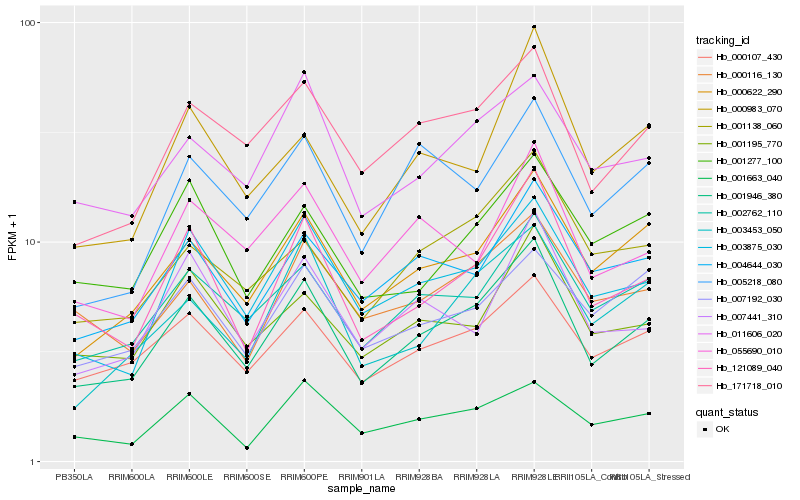
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001946_380 |
0.0 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 34-like [Jatropha curcas] |
| 2 |
Hb_121089_040 |
0.0783671597 |
- |
- |
PREDICTED: GDSL esterase/lipase At4g10955 [Jatropha curcas] |
| 3 |
Hb_000622_290 |
0.0835136553 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At3g61590 [Jatropha curcas] |
| 4 |
Hb_002762_110 |
0.0835967797 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_171718_010 |
0.085012314 |
- |
- |
PREDICTED: disease resistance protein At4g27190-like [Eucalyptus grandis] |
| 6 |
Hb_000107_430 |
0.0868334388 |
- |
- |
PREDICTED: brefeldin A-inhibited guanine nucleotide-exchange protein 1 [Jatropha curcas] |
| 7 |
Hb_003875_030 |
0.0869145393 |
- |
- |
PREDICTED: probable plastidic glucose transporter 1 [Jatropha curcas] |
| 8 |
Hb_004644_030 |
0.0997429681 |
- |
- |
PREDICTED: FAD synthase isoform X3 [Jatropha curcas] |
| 9 |
Hb_011606_020 |
0.1022261726 |
- |
- |
hypothetical protein CISIN_1g023687mg [Citrus sinensis] |
| 10 |
Hb_001195_770 |
0.1024380259 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase ARI2 [Jatropha curcas] |
| 11 |
Hb_001138_060 |
0.1046232396 |
- |
- |
hypothetical protein CICLE_v10028086mg [Citrus clementina] |
| 12 |
Hb_003453_050 |
0.1049661361 |
- |
- |
PREDICTED: calcium-binding mitochondrial carrier protein SCaMC-3-like [Jatropha curcas] |
| 13 |
Hb_001663_040 |
0.1085869227 |
- |
- |
hypothetical protein B456_007G162000 [Gossypium raimondii] |
| 14 |
Hb_000116_130 |
0.1106695624 |
- |
- |
PREDICTED: aspartic proteinase Asp1 [Jatropha curcas] |
| 15 |
Hb_055690_010 |
0.1107068201 |
- |
- |
PREDICTED: pheophytinase, chloroplastic [Jatropha curcas] |
| 16 |
Hb_007441_310 |
0.1116379235 |
- |
- |
PREDICTED: protein TRIGALACTOSYLDIACYLGLYCEROL 4, chloroplastic [Jatropha curcas] |
| 17 |
Hb_001277_100 |
0.1119065833 |
- |
- |
PREDICTED: 2-methyl-6-phytyl-1,4-hydroquinone methyltransferase, chloroplastic-like isoform X1 [Gossypium raimondii] |
| 18 |
Hb_005218_080 |
0.1137791946 |
- |
- |
PREDICTED: branched-chain-amino-acid aminotransferase 3, chloroplastic isoform X1 [Jatropha curcas] |
| 19 |
Hb_000983_070 |
0.1150251745 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_007192_030 |
0.1162248457 |
- |
- |
PREDICTED: manganese-dependent ADP-ribose/CDP-alcohol diphosphatase [Jatropha curcas] |