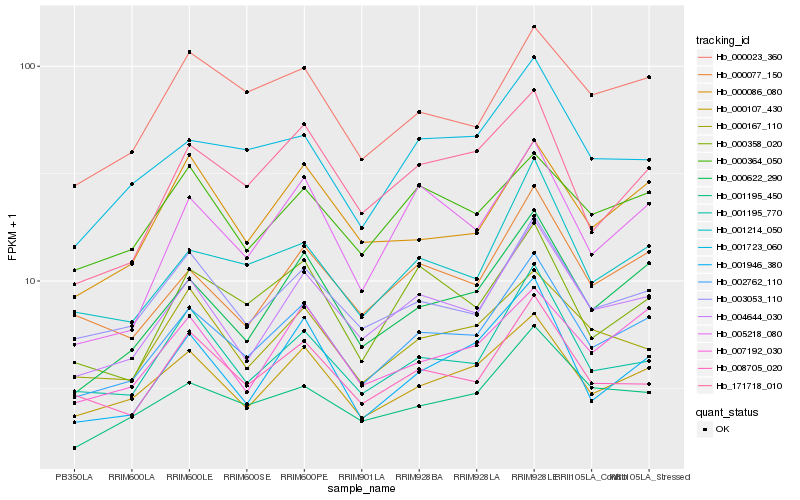
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002762_110 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_004644_030 |
0.0548459788 |
- |
- |
PREDICTED: FAD synthase isoform X3 [Jatropha curcas] |
| 3 |
Hb_000622_290 |
0.0581437363 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At3g61590 [Jatropha curcas] |
| 4 |
Hb_000107_430 |
0.0605525325 |
- |
- |
PREDICTED: brefeldin A-inhibited guanine nucleotide-exchange protein 1 [Jatropha curcas] |
| 5 |
Hb_003053_110 |
0.0682842589 |
- |
- |
PREDICTED: protease Do-like 1, chloroplastic [Jatropha curcas] |
| 6 |
Hb_000077_150 |
0.0699228177 |
- |
- |
PREDICTED: riboflavin synthase [Jatropha curcas] |
| 7 |
Hb_001214_050 |
0.0706848253 |
- |
- |
PREDICTED: bifunctional monothiol glutaredoxin-S16, chloroplastic [Jatropha curcas] |
| 8 |
Hb_001195_770 |
0.0753184986 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase ARI2 [Jatropha curcas] |
| 9 |
Hb_001195_450 |
0.0757539127 |
- |
- |
PREDICTED: uncharacterized protein LOC105633771 [Jatropha curcas] |
| 10 |
Hb_008705_020 |
0.0814802973 |
- |
- |
PREDICTED: lisH domain and HEAT repeat-containing protein KIAA1468 homolog isoform X1 [Jatropha curcas] |
| 11 |
Hb_000023_360 |
0.0817201681 |
- |
- |
phospholipid hydroperoxide glutathione peroxidase 1, chloroplastic [Jatropha curcas] |
| 12 |
Hb_001723_060 |
0.0822832178 |
rubber biosynthesis |
Gene Name: Dihydrolipoamide dehydrogenase |
PREDICTED: LOW QUALITY PROTEIN: dihydrolipoyl dehydrogenase 2, mitochondrial [Populus euphratica] |
| 13 |
Hb_007192_030 |
0.0835633016 |
- |
- |
PREDICTED: manganese-dependent ADP-ribose/CDP-alcohol diphosphatase [Jatropha curcas] |
| 14 |
Hb_001946_380 |
0.0835967797 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 34-like [Jatropha curcas] |
| 15 |
Hb_000358_020 |
0.0849254212 |
- |
- |
PREDICTED: uncharacterized protein C05D11.7-like [Jatropha curcas] |
| 16 |
Hb_171718_010 |
0.0850107769 |
- |
- |
PREDICTED: disease resistance protein At4g27190-like [Eucalyptus grandis] |
| 17 |
Hb_000086_080 |
0.0887151687 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000167_110 |
0.0889961672 |
- |
- |
glycerol-3-phosphate dehydrogenase, putative [Ricinus communis] |
| 19 |
Hb_005218_080 |
0.0896912519 |
- |
- |
PREDICTED: branched-chain-amino-acid aminotransferase 3, chloroplastic isoform X1 [Jatropha curcas] |
| 20 |
Hb_000364_050 |
0.0900408341 |
- |
- |
PREDICTED: CBS domain-containing protein CBSX1, chloroplastic [Jatropha curcas] |