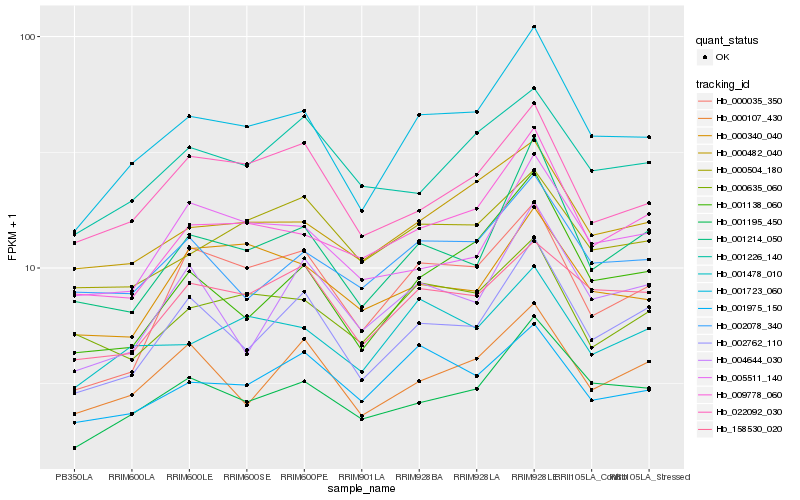
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001723_060 |
0.0 |
rubber biosynthesis |
Gene Name: Dihydrolipoamide dehydrogenase |
PREDICTED: LOW QUALITY PROTEIN: dihydrolipoyl dehydrogenase 2, mitochondrial [Populus euphratica] |
| 2 |
Hb_001195_450 |
0.0682053234 |
- |
- |
PREDICTED: uncharacterized protein LOC105633771 [Jatropha curcas] |
| 3 |
Hb_002762_110 |
0.0822832178 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_009778_060 |
0.0848855147 |
- |
- |
PREDICTED: uncharacterized protein LOC105636987 [Jatropha curcas] |
| 5 |
Hb_001138_060 |
0.0864781268 |
- |
- |
hypothetical protein CICLE_v10028086mg [Citrus clementina] |
| 6 |
Hb_001214_050 |
0.0869576296 |
- |
- |
PREDICTED: bifunctional monothiol glutaredoxin-S16, chloroplastic [Jatropha curcas] |
| 7 |
Hb_000482_040 |
0.0890439185 |
- |
- |
Protein YME1, putative [Ricinus communis] |
| 8 |
Hb_001478_010 |
0.0948880874 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g48250, chloroplastic [Jatropha curcas] |
| 9 |
Hb_158530_020 |
0.0981871335 |
- |
- |
PREDICTED: aspartic proteinase CDR1 [Jatropha curcas] |
| 10 |
Hb_000035_350 |
0.0987884812 |
- |
- |
PREDICTED: monogalactosyldiacylglycerol synthase, chloroplastic [Jatropha curcas] |
| 11 |
Hb_002078_340 |
0.0997303645 |
- |
- |
PREDICTED: uncharacterized protein LOC105644820 isoform X2 [Jatropha curcas] |
| 12 |
Hb_022092_030 |
0.0999949956 |
- |
- |
PREDICTED: lipoyl synthase, mitochondrial [Jatropha curcas] |
| 13 |
Hb_000107_430 |
0.1039249661 |
- |
- |
PREDICTED: brefeldin A-inhibited guanine nucleotide-exchange protein 1 [Jatropha curcas] |
| 14 |
Hb_005511_140 |
0.1048333932 |
- |
- |
Ubiquinone biosynthesis protein coq-8, putative [Ricinus communis] |
| 15 |
Hb_001975_150 |
0.1065228765 |
- |
- |
PREDICTED: RINT1-like protein MAG2L [Jatropha curcas] |
| 16 |
Hb_001226_140 |
0.1093472317 |
- |
- |
PREDICTED: protein phosphatase 2C 77-like [Jatropha curcas] |
| 17 |
Hb_000635_060 |
0.1095621316 |
- |
- |
PREDICTED: uncharacterized protein LOC105644797 [Jatropha curcas] |
| 18 |
Hb_004644_030 |
0.1099031838 |
- |
- |
PREDICTED: FAD synthase isoform X3 [Jatropha curcas] |
| 19 |
Hb_000504_180 |
0.110065976 |
- |
- |
PREDICTED: heterogeneous nuclear ribonucleoprotein 1 [Jatropha curcas] |
| 20 |
Hb_000340_040 |
0.110995518 |
- |
- |
PREDICTED: HUA2-like protein 3 isoform X3 [Jatropha curcas] |