| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
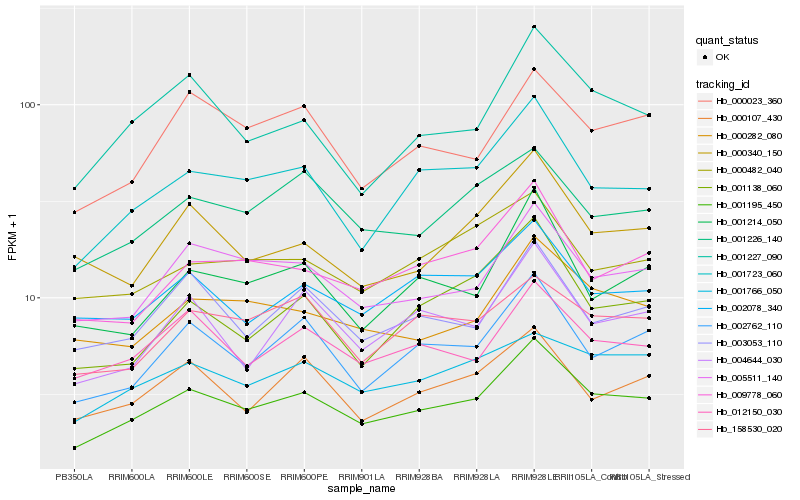
Hb_001195_450 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105633771 [Jatropha curcas] |
| 2 |
Hb_001723_060 |
0.0682053234 |
rubber biosynthesis |
Gene Name: Dihydrolipoamide dehydrogenase |
PREDICTED: LOW QUALITY PROTEIN: dihydrolipoyl dehydrogenase 2, mitochondrial [Populus euphratica] |
| 3 |
Hb_002762_110 |
0.0757539127 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_005511_140 |
0.0776457227 |
- |
- |
Ubiquinone biosynthesis protein coq-8, putative [Ricinus communis] |
| 5 |
Hb_009778_060 |
0.0837537141 |
- |
- |
PREDICTED: uncharacterized protein LOC105636987 [Jatropha curcas] |
| 6 |
Hb_012150_030 |
0.0838599784 |
- |
- |
PREDICTED: aspartate--tRNA ligase, mitochondrial [Jatropha curcas] |
| 7 |
Hb_000282_080 |
0.0843206115 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_001227_090 |
0.0854815103 |
- |
- |
peroxiredoxin, putative [Ricinus communis] |
| 9 |
Hb_003053_110 |
0.087871786 |
- |
- |
PREDICTED: protease Do-like 1, chloroplastic [Jatropha curcas] |
| 10 |
Hb_001226_140 |
0.0881055818 |
- |
- |
PREDICTED: protein phosphatase 2C 77-like [Jatropha curcas] |
| 11 |
Hb_001766_050 |
0.0891338868 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g29290 [Jatropha curcas] |
| 12 |
Hb_002078_340 |
0.089888395 |
- |
- |
PREDICTED: uncharacterized protein LOC105644820 isoform X2 [Jatropha curcas] |
| 13 |
Hb_004644_030 |
0.0899051952 |
- |
- |
PREDICTED: FAD synthase isoform X3 [Jatropha curcas] |
| 14 |
Hb_000023_360 |
0.0917205145 |
- |
- |
phospholipid hydroperoxide glutathione peroxidase 1, chloroplastic [Jatropha curcas] |
| 15 |
Hb_000107_430 |
0.0917222908 |
- |
- |
PREDICTED: brefeldin A-inhibited guanine nucleotide-exchange protein 1 [Jatropha curcas] |
| 16 |
Hb_001214_050 |
0.092084121 |
- |
- |
PREDICTED: bifunctional monothiol glutaredoxin-S16, chloroplastic [Jatropha curcas] |
| 17 |
Hb_158530_020 |
0.0943871794 |
- |
- |
PREDICTED: aspartic proteinase CDR1 [Jatropha curcas] |
| 18 |
Hb_001138_060 |
0.0950181378 |
- |
- |
hypothetical protein CICLE_v10028086mg [Citrus clementina] |
| 19 |
Hb_000482_040 |
0.0972111214 |
- |
- |
Protein YME1, putative [Ricinus communis] |
| 20 |
Hb_000340_150 |
0.0996677921 |
- |
- |
hypothetical protein JCGZ_21975 [Jatropha curcas] |