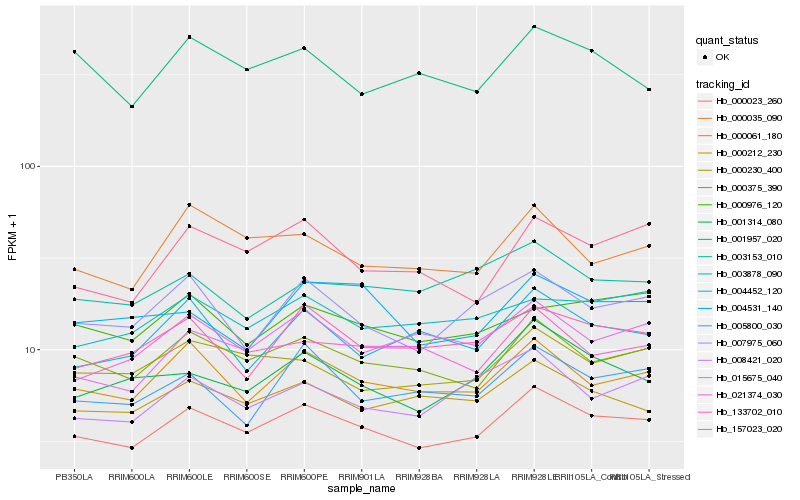
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000023_260 |
0.0 |
- |
- |
PREDICTED: monogalactosyldiacylglycerol synthase, chloroplastic [Jatropha curcas] |
| 2 |
Hb_000375_390 |
0.0566327702 |
- |
- |
poly-A binding protein, putative [Ricinus communis] |
| 3 |
Hb_000061_180 |
0.0611510774 |
- |
- |
exonuclease, putative [Ricinus communis] |
| 4 |
Hb_000212_230 |
0.0719112218 |
- |
- |
PREDICTED: endoribonuclease Dicer homolog 2 [Jatropha curcas] |
| 5 |
Hb_001314_080 |
0.0750825702 |
- |
- |
hypothetical protein POPTR_0018s14330g [Populus trichocarpa] |
| 6 |
Hb_133702_010 |
0.0761592147 |
- |
- |
PREDICTED: reticulon-4-interacting protein 1, mitochondrial isoform X1 [Jatropha curcas] |
| 7 |
Hb_007975_060 |
0.0762202282 |
- |
- |
PREDICTED: uncharacterized GPI-anchored protein At1g61900 isoform X2 [Jatropha curcas] |
| 8 |
Hb_000230_400 |
0.0784851513 |
- |
- |
PREDICTED: protein ABCI12, chloroplastic isoform X1 [Jatropha curcas] |
| 9 |
Hb_000035_090 |
0.0807484038 |
- |
- |
PREDICTED: stromal cell-derived factor 2-like protein [Jatropha curcas] |
| 10 |
Hb_008421_020 |
0.080760451 |
- |
- |
PREDICTED: uncharacterized protein LOC105635546 isoform X2 [Jatropha curcas] |
| 11 |
Hb_157023_020 |
0.0829135523 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 12 |
Hb_005800_030 |
0.0840211276 |
- |
- |
PREDICTED: lysM and putative peptidoglycan-binding domain-containing protein 4-like [Jatropha curcas] |
| 13 |
Hb_021374_030 |
0.0852100933 |
- |
- |
hypothetical protein RCOM_0351490 [Ricinus communis] |
| 14 |
Hb_003878_090 |
0.0860645056 |
- |
- |
PREDICTED: polynucleotide 3'-phosphatase ZDP [Jatropha curcas] |
| 15 |
Hb_004452_120 |
0.0865243717 |
- |
- |
PREDICTED: uncharacterized protein LOC105639574 [Jatropha curcas] |
| 16 |
Hb_015675_040 |
0.087131844 |
- |
- |
PREDICTED: ribose-phosphate pyrophosphokinase 1 [Jatropha curcas] |
| 17 |
Hb_003153_010 |
0.087367717 |
- |
- |
PREDICTED: diacylglycerol kinase 3-like [Jatropha curcas] |
| 18 |
Hb_001957_020 |
0.088867836 |
- |
- |
glutathione-S-transferase tau 1 [Hevea brasiliensis] |
| 19 |
Hb_000976_120 |
0.0897807024 |
- |
- |
PREDICTED: DNA polymerase zeta processivity subunit [Jatropha curcas] |
| 20 |
Hb_004531_140 |
0.0901014137 |
- |
- |
PREDICTED: uncharacterized protein LOC105635023 [Jatropha curcas] |