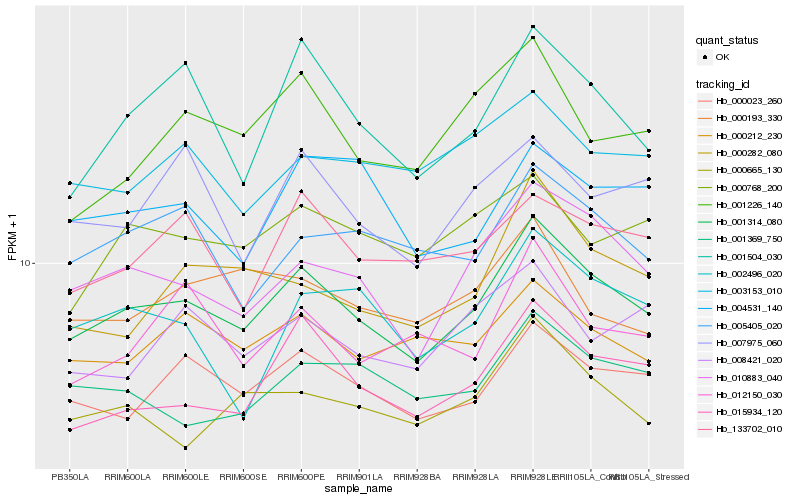
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001314_080 |
0.0 |
- |
- |
hypothetical protein POPTR_0018s14330g [Populus trichocarpa] |
| 2 |
Hb_010883_040 |
0.068568748 |
- |
- |
PREDICTED: phosphatidylinositol 4-phosphate 5-kinase 1 [Jatropha curcas] |
| 3 |
Hb_000023_260 |
0.0750825702 |
- |
- |
PREDICTED: monogalactosyldiacylglycerol synthase, chloroplastic [Jatropha curcas] |
| 4 |
Hb_015934_120 |
0.0827040379 |
- |
- |
PREDICTED: callose synthase 7 [Vitis vinifera] |
| 5 |
Hb_001504_030 |
0.088684399 |
- |
- |
PREDICTED: probable CCR4-associated factor 1 homolog 6 [Jatropha curcas] |
| 6 |
Hb_000212_230 |
0.0913020574 |
- |
- |
PREDICTED: endoribonuclease Dicer homolog 2 [Jatropha curcas] |
| 7 |
Hb_000665_130 |
0.0944872503 |
- |
- |
PREDICTED: uncharacterized protein LOC105637599 isoform X2 [Jatropha curcas] |
| 8 |
Hb_005405_020 |
0.0945677555 |
- |
- |
PREDICTED: petal death protein isoform X1 [Jatropha curcas] |
| 9 |
Hb_133702_010 |
0.0948033265 |
- |
- |
PREDICTED: reticulon-4-interacting protein 1, mitochondrial isoform X1 [Jatropha curcas] |
| 10 |
Hb_000282_080 |
0.0950331409 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_001369_750 |
0.0956496777 |
- |
- |
PREDICTED: probable rRNA-processing protein EBP2 homolog [Jatropha curcas] |
| 12 |
Hb_003153_010 |
0.0972974895 |
- |
- |
PREDICTED: diacylglycerol kinase 3-like [Jatropha curcas] |
| 13 |
Hb_001226_140 |
0.0978202427 |
- |
- |
PREDICTED: protein phosphatase 2C 77-like [Jatropha curcas] |
| 14 |
Hb_000768_200 |
0.0979166255 |
- |
- |
hypothetical protein POPTR_0013s06910g [Populus trichocarpa] |
| 15 |
Hb_002496_020 |
0.0993847265 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 16 |
Hb_007975_060 |
0.1006117353 |
- |
- |
PREDICTED: uncharacterized GPI-anchored protein At1g61900 isoform X2 [Jatropha curcas] |
| 17 |
Hb_008421_020 |
0.1010072822 |
- |
- |
PREDICTED: uncharacterized protein LOC105635546 isoform X2 [Jatropha curcas] |
| 18 |
Hb_000193_330 |
0.1018577144 |
- |
- |
PREDICTED: formin-like protein 20 [Jatropha curcas] |
| 19 |
Hb_004531_140 |
0.1024583715 |
- |
- |
PREDICTED: uncharacterized protein LOC105635023 [Jatropha curcas] |
| 20 |
Hb_012150_030 |
0.1027891866 |
- |
- |
PREDICTED: aspartate--tRNA ligase, mitochondrial [Jatropha curcas] |