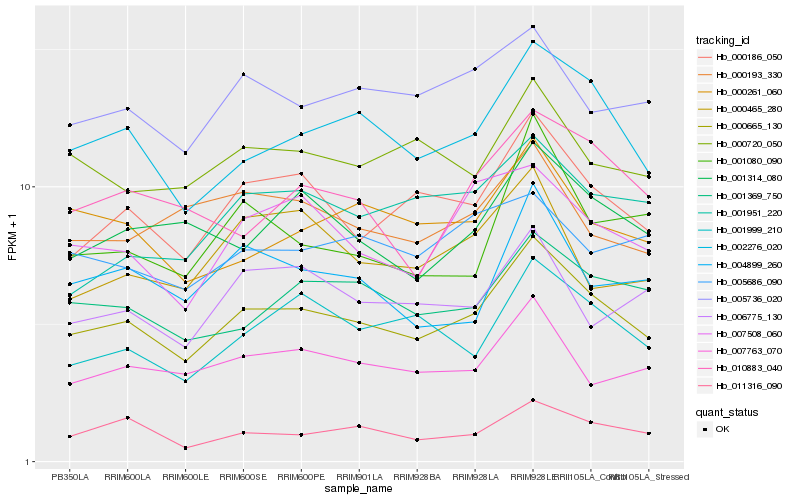
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000665_130 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105637599 isoform X2 [Jatropha curcas] |
| 2 |
Hb_002276_020 |
0.0667295748 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH49 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000186_050 |
0.0728809041 |
- |
- |
PREDICTED: uncharacterized protein LOC105649281 [Jatropha curcas] |
| 4 |
Hb_001369_750 |
0.0867921852 |
- |
- |
PREDICTED: probable rRNA-processing protein EBP2 homolog [Jatropha curcas] |
| 5 |
Hb_011316_090 |
0.0886213336 |
- |
- |
OSJNBa0004L19.22 [Oryza sativa Japonica Group] |
| 6 |
Hb_005736_020 |
0.0914463351 |
- |
- |
hypothetical protein POPTR_0005s20940g [Populus trichocarpa] |
| 7 |
Hb_007508_060 |
0.0934383039 |
- |
- |
PREDICTED: uncharacterized protein LOC105640628 [Jatropha curcas] |
| 8 |
Hb_000261_060 |
0.0939861014 |
- |
- |
PREDICTED: pre-mRNA-splicing factor 38B-like [Jatropha curcas] |
| 9 |
Hb_001314_080 |
0.0944872503 |
- |
- |
hypothetical protein POPTR_0018s14330g [Populus trichocarpa] |
| 10 |
Hb_007763_070 |
0.0944872728 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g71460, chloroplastic [Jatropha curcas] |
| 11 |
Hb_000720_050 |
0.0954105526 |
- |
- |
PREDICTED: uncharacterized protein LOC105645857 isoform X1 [Jatropha curcas] |
| 12 |
Hb_001999_210 |
0.0979481197 |
- |
- |
PREDICTED: sodium-dependent phosphate transport protein 1, chloroplastic [Eucalyptus grandis] |
| 13 |
Hb_004899_260 |
0.1016930693 |
- |
- |
PREDICTED: uncharacterized protein LOC105638006 [Jatropha curcas] |
| 14 |
Hb_000193_330 |
0.103416981 |
- |
- |
PREDICTED: formin-like protein 20 [Jatropha curcas] |
| 15 |
Hb_010883_040 |
0.1038831647 |
- |
- |
PREDICTED: phosphatidylinositol 4-phosphate 5-kinase 1 [Jatropha curcas] |
| 16 |
Hb_006775_130 |
0.1046343753 |
- |
- |
PREDICTED: bidirectional sugar transporter SWEET2a-like [Jatropha curcas] |
| 17 |
Hb_001080_090 |
0.1058428574 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At2g30600 isoform X1 [Jatropha curcas] |
| 18 |
Hb_005686_090 |
0.1062695223 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 2 isoform X1 [Jatropha curcas] |
| 19 |
Hb_001951_220 |
0.1068998278 |
- |
- |
PREDICTED: CTD small phosphatase-like protein 2 isoform X5 [Jatropha curcas] |
| 20 |
Hb_000465_280 |
0.1072877668 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g47360 [Jatropha curcas] |