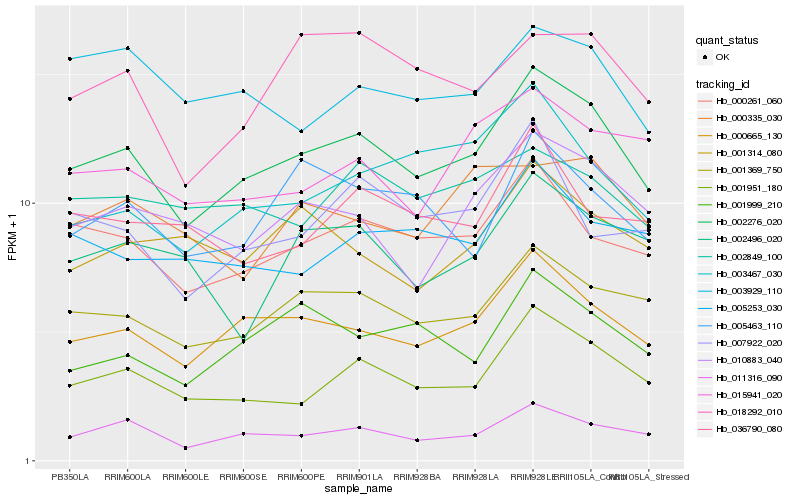
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002276_020 |
0.0 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH49 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000665_130 |
0.0667295748 |
- |
- |
PREDICTED: uncharacterized protein LOC105637599 isoform X2 [Jatropha curcas] |
| 3 |
Hb_011316_090 |
0.0750395165 |
- |
- |
OSJNBa0004L19.22 [Oryza sativa Japonica Group] |
| 4 |
Hb_001369_750 |
0.0775751249 |
- |
- |
PREDICTED: probable rRNA-processing protein EBP2 homolog [Jatropha curcas] |
| 5 |
Hb_001951_180 |
0.0826194562 |
- |
- |
PREDICTED: carboxyl-terminal-processing peptidase 1, chloroplastic isoform X2 [Jatropha curcas] |
| 6 |
Hb_000261_060 |
0.0829005871 |
- |
- |
PREDICTED: pre-mRNA-splicing factor 38B-like [Jatropha curcas] |
| 7 |
Hb_010883_040 |
0.1009924731 |
- |
- |
PREDICTED: phosphatidylinositol 4-phosphate 5-kinase 1 [Jatropha curcas] |
| 8 |
Hb_015941_020 |
0.1085202067 |
- |
- |
- |
| 9 |
Hb_005253_030 |
0.1086168976 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 10 |
Hb_018292_010 |
0.108846568 |
transcription factor |
TF Family: NF-YB |
PREDICTED: nuclear transcription factor Y subunit B-3 [Jatropha curcas] |
| 11 |
Hb_001999_210 |
0.1094502236 |
- |
- |
PREDICTED: sodium-dependent phosphate transport protein 1, chloroplastic [Eucalyptus grandis] |
| 12 |
Hb_005463_110 |
0.1095318075 |
- |
- |
PREDICTED: glyoxylate/hydroxypyruvate reductase HPR3-like [Jatropha curcas] |
| 13 |
Hb_002849_100 |
0.1100356221 |
- |
- |
PREDICTED: mechanosensitive ion channel protein 1, mitochondrial [Jatropha curcas] |
| 14 |
Hb_003467_030 |
0.1108873673 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At5g02620-like [Jatropha curcas] |
| 15 |
Hb_007922_020 |
0.1129064232 |
- |
- |
PREDICTED: uncharacterized protein LOC105646505 [Jatropha curcas] |
| 16 |
Hb_036790_080 |
0.11509127 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 39 [Jatropha curcas] |
| 17 |
Hb_003929_110 |
0.1161571391 |
- |
- |
PREDICTED: probable GTP diphosphokinase CRSH, chloroplastic isoform X1 [Jatropha curcas] |
| 18 |
Hb_001314_080 |
0.1162522978 |
- |
- |
hypothetical protein POPTR_0018s14330g [Populus trichocarpa] |
| 19 |
Hb_002496_020 |
0.1173244488 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 20 |
Hb_000335_030 |
0.1189153728 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g17033 [Jatropha curcas] |