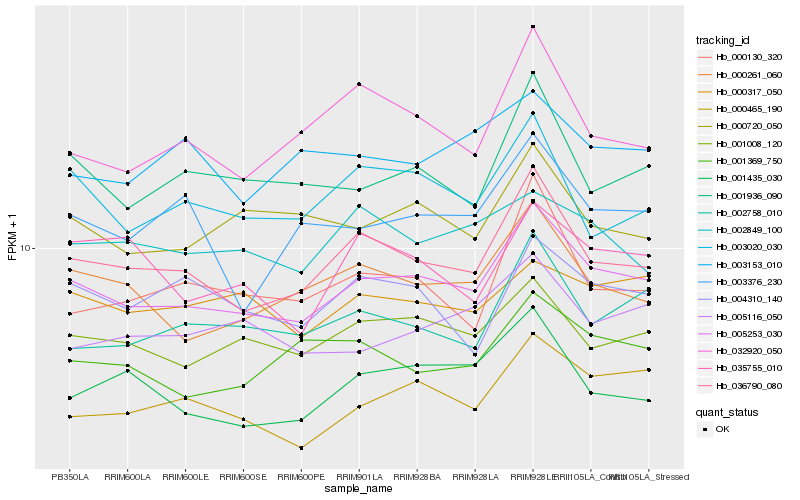
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005253_030 |
0.0 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 2 |
Hb_036790_080 |
0.0556495721 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 39 [Jatropha curcas] |
| 3 |
Hb_001008_120 |
0.0698034104 |
desease resistance |
Gene Name: PEX-1N |
peroxisome biogenesis factor, putative [Ricinus communis] |
| 4 |
Hb_000261_060 |
0.0734468668 |
- |
- |
PREDICTED: pre-mRNA-splicing factor 38B-like [Jatropha curcas] |
| 5 |
Hb_001936_090 |
0.0809951933 |
- |
- |
PREDICTED: BOI-related E3 ubiquitin-protein ligase 1-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_000465_190 |
0.081331726 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase HOS1 isoform X2 [Jatropha curcas] |
| 7 |
Hb_002758_010 |
0.0848790288 |
- |
- |
PREDICTED: ribonuclease II, chloroplastic/mitochondrial [Populus euphratica] |
| 8 |
Hb_000720_050 |
0.0849048757 |
- |
- |
PREDICTED: uncharacterized protein LOC105645857 isoform X1 [Jatropha curcas] |
| 9 |
Hb_003020_030 |
0.0867138518 |
- |
- |
PREDICTED: protein TOC75-3, chloroplastic-like [Populus euphratica] |
| 10 |
Hb_005116_050 |
0.0879819636 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g02820, mitochondrial isoform X1 [Jatropha curcas] |
| 11 |
Hb_001369_750 |
0.0881602694 |
- |
- |
PREDICTED: probable rRNA-processing protein EBP2 homolog [Jatropha curcas] |
| 12 |
Hb_000317_050 |
0.0889143493 |
- |
- |
formiminotransferase-cyclodeaminase, putative [Ricinus communis] |
| 13 |
Hb_032920_050 |
0.0899205485 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit-related protein 1, chloroplastic [Jatropha curcas] |
| 14 |
Hb_004310_140 |
0.0909672863 |
- |
- |
PREDICTED: glycine--tRNA ligase 2, chloroplastic/mitochondrial isoform X1 [Jatropha curcas] |
| 15 |
Hb_003376_230 |
0.0918543735 |
- |
- |
PREDICTED: dnaJ protein P58IPK homolog [Jatropha curcas] |
| 16 |
Hb_002849_100 |
0.093732332 |
- |
- |
PREDICTED: mechanosensitive ion channel protein 1, mitochondrial [Jatropha curcas] |
| 17 |
Hb_000130_320 |
0.0941620523 |
desease resistance |
Gene Name: AAA |
PREDICTED: uncharacterized protein ycf45 [Jatropha curcas] |
| 18 |
Hb_001435_030 |
0.0949004087 |
- |
- |
PREDICTED: uncharacterized protein LOC105645073 [Jatropha curcas] |
| 19 |
Hb_003153_010 |
0.0950122775 |
- |
- |
PREDICTED: diacylglycerol kinase 3-like [Jatropha curcas] |
| 20 |
Hb_035755_010 |
0.0955218524 |
- |
- |
unknown [Populus trichocarpa] |