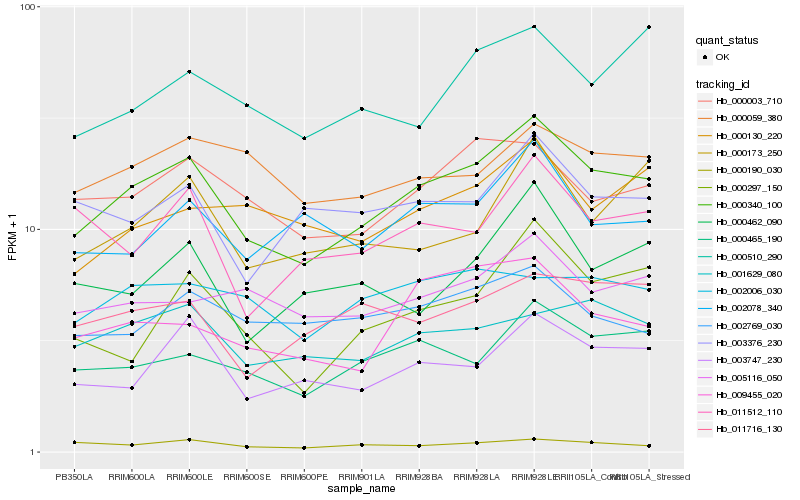
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000340_100 |
0.0 |
- |
- |
PREDICTED: protein PALE CRESS, chloroplastic [Jatropha curcas] |
| 2 |
Hb_000465_190 |
0.1036151023 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase HOS1 isoform X2 [Jatropha curcas] |
| 3 |
Hb_000003_710 |
0.1090449478 |
- |
- |
PREDICTED: RNA polymerase II-associated factor 1 homolog [Jatropha curcas] |
| 4 |
Hb_003747_230 |
0.1096210437 |
- |
- |
PREDICTED: uncharacterized protein LOC105630239 isoform X1 [Jatropha curcas] |
| 5 |
Hb_001629_080 |
0.1146396616 |
- |
- |
hypothetical protein POPTR_0011s12610g [Populus trichocarpa] |
| 6 |
Hb_005116_050 |
0.1174617921 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g02820, mitochondrial isoform X1 [Jatropha curcas] |
| 7 |
Hb_002078_340 |
0.1202604549 |
- |
- |
PREDICTED: uncharacterized protein LOC105644820 isoform X2 [Jatropha curcas] |
| 8 |
Hb_009455_020 |
0.1221253962 |
- |
- |
PREDICTED: uncharacterized protein LOC105634581 [Jatropha curcas] |
| 9 |
Hb_011716_130 |
0.1225401282 |
- |
- |
PREDICTED: probable acyl-[acyl-carrier-protein]--UDP-N-acetylglucosamine O-acyltransferase, mitochondrial isoform X5 [Populus euphratica] |
| 10 |
Hb_000510_290 |
0.1225695106 |
- |
- |
PREDICTED: protein OSB2, chloroplastic-like isoform X2 [Jatropha curcas] |
| 11 |
Hb_000190_030 |
0.1227524591 |
- |
- |
PREDICTED: BEACH domain-containing protein lvsA-like [Pyrus x bretschneideri] |
| 12 |
Hb_000297_150 |
0.1230822678 |
- |
- |
PREDICTED: uncharacterized protein LOC103500908 [Cucumis melo] |
| 13 |
Hb_003376_230 |
0.1234974852 |
- |
- |
PREDICTED: dnaJ protein P58IPK homolog [Jatropha curcas] |
| 14 |
Hb_002769_030 |
0.1236533191 |
- |
- |
PREDICTED: MATE efflux family protein 4, chloroplastic-like [Jatropha curcas] |
| 15 |
Hb_000059_380 |
0.1239777034 |
- |
- |
PREDICTED: uncharacterized protein LOC105648668 [Jatropha curcas] |
| 16 |
Hb_000173_250 |
0.1241414271 |
- |
- |
PREDICTED: ribonucleoside-diphosphate reductase small chain A [Jatropha curcas] |
| 17 |
Hb_000130_220 |
0.1246957711 |
- |
- |
PREDICTED: uncharacterized protein At3g15000, mitochondrial-like [Jatropha curcas] |
| 18 |
Hb_000462_090 |
0.1254574269 |
- |
- |
PREDICTED: carotene epsilon-monooxygenase, chloroplastic [Jatropha curcas] |
| 19 |
Hb_002006_030 |
0.1256279696 |
- |
- |
PREDICTED: zinc finger BED domain-containing protein DAYSLEEPER [Jatropha curcas] |
| 20 |
Hb_011512_110 |
0.1263139265 |
- |
- |
PREDICTED: GDT1-like protein 2, chloroplastic isoform X1 [Jatropha curcas] |