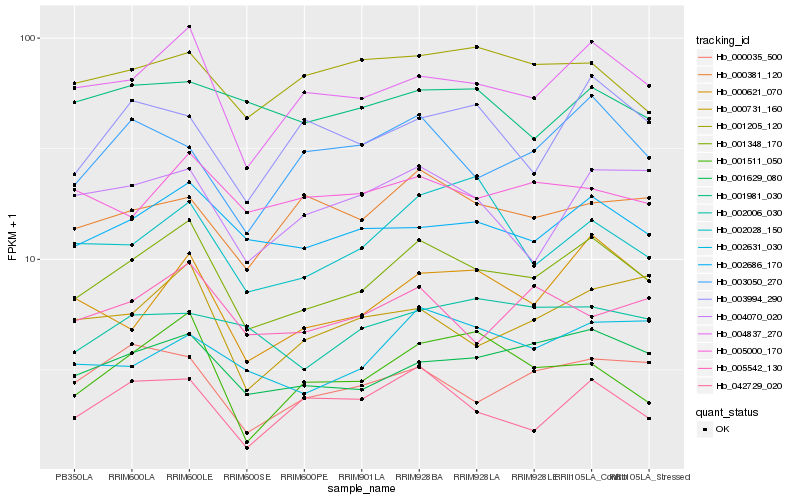
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001348_170 |
0.0 |
- |
- |
PREDICTED: filament-like plant protein isoform X2 [Jatropha curcas] |
| 2 |
Hb_004837_270 |
0.0709097857 |
- |
- |
PREDICTED: cinnamoyl-CoA reductase 1 [Jatropha curcas] |
| 3 |
Hb_001629_080 |
0.0865099237 |
- |
- |
hypothetical protein POPTR_0011s12610g [Populus trichocarpa] |
| 4 |
Hb_002686_170 |
0.0885953592 |
- |
- |
PREDICTED: uncharacterized protein LOC105632645 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000621_070 |
0.1052960673 |
- |
- |
PREDICTED: caffeoylshikimate esterase [Jatropha curcas] |
| 6 |
Hb_000035_500 |
0.1133654157 |
- |
- |
PREDICTED: ADP-ribosylation factor 1 isoform X2 [Tarenaya hassleriana] |
| 7 |
Hb_004070_020 |
0.1139858655 |
- |
- |
rhythmically-expressed protein 2 protein, putative [Ricinus communis] |
| 8 |
Hb_005542_130 |
0.1149089119 |
- |
- |
PREDICTED: uncharacterized protein LOC105649174 [Jatropha curcas] |
| 9 |
Hb_000731_160 |
0.1168538966 |
- |
- |
PREDICTED: adenosine deaminase-like protein [Jatropha curcas] |
| 10 |
Hb_000381_120 |
0.1177793709 |
- |
- |
PREDICTED: uncharacterized protein LOC105648175 [Jatropha curcas] |
| 11 |
Hb_003050_270 |
0.1178366177 |
- |
- |
PREDICTED: 3-oxoacyl-[acyl-carrier-protein] synthase I, chloroplastic [Jatropha curcas] |
| 12 |
Hb_042729_020 |
0.1182236075 |
- |
- |
PREDICTED: protein argonaute 16 isoform X1 [Jatropha curcas] |
| 13 |
Hb_005000_170 |
0.1185888967 |
- |
- |
PREDICTED: WD-40 repeat-containing protein MSI4 [Jatropha curcas] |
| 14 |
Hb_001205_120 |
0.1188828048 |
- |
- |
PREDICTED: succinate dehydrogenase [ubiquinone] flavoprotein subunit 1, mitochondrial [Populus euphratica] |
| 15 |
Hb_002631_030 |
0.1190348857 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_002028_150 |
0.1194950139 |
- |
- |
PREDICTED: hepatocyte growth factor-regulated tyrosine kinase substrate [Jatropha curcas] |
| 17 |
Hb_001511_050 |
0.1216171695 |
- |
- |
PREDICTED: probable inactive serine/threonine-protein kinase bub1 isoform X1 [Jatropha curcas] |
| 18 |
Hb_003994_290 |
0.1237500043 |
- |
- |
PREDICTED: glyceraldehyde-3-phosphate dehydrogenase GAPCP2, chloroplastic [Jatropha curcas] |
| 19 |
Hb_002006_030 |
0.1238386017 |
- |
- |
PREDICTED: zinc finger BED domain-containing protein DAYSLEEPER [Jatropha curcas] |
| 20 |
Hb_001981_030 |
0.1247040407 |
- |
- |
nuclear acid binding protein, putative [Ricinus communis] |