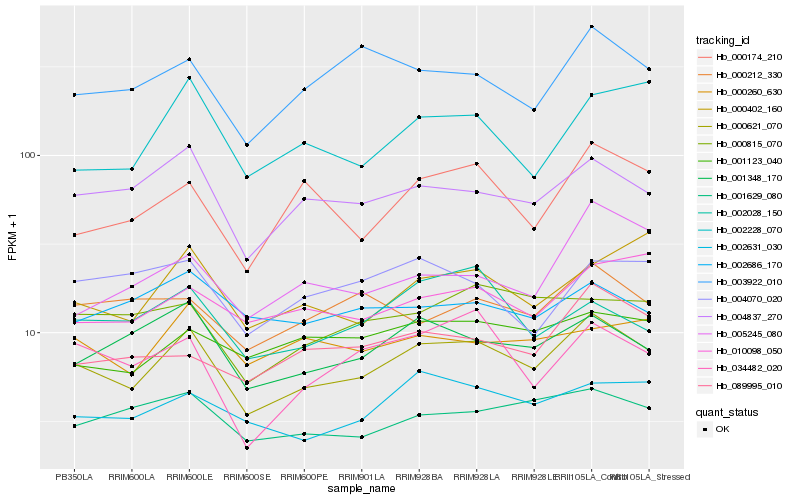
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000621_070 |
0.0 |
- |
- |
PREDICTED: caffeoylshikimate esterase [Jatropha curcas] |
| 2 |
Hb_004837_270 |
0.0955251437 |
- |
- |
PREDICTED: cinnamoyl-CoA reductase 1 [Jatropha curcas] |
| 3 |
Hb_001348_170 |
0.1052960673 |
- |
- |
PREDICTED: filament-like plant protein isoform X2 [Jatropha curcas] |
| 4 |
Hb_002631_030 |
0.1102185012 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_001629_080 |
0.1118794106 |
- |
- |
hypothetical protein POPTR_0011s12610g [Populus trichocarpa] |
| 6 |
Hb_000174_210 |
0.1157357153 |
- |
- |
latex cyanogenic beta glucosidase [Hevea brasiliensis] |
| 7 |
Hb_034482_020 |
0.1167258509 |
- |
- |
PREDICTED: bifunctional aspartokinase/homoserine dehydrogenase 1, chloroplastic-like [Jatropha curcas] |
| 8 |
Hb_003922_010 |
0.1167387774 |
- |
- |
PREDICTED: probable phospholipid hydroperoxide glutathione peroxidase [Jatropha curcas] |
| 9 |
Hb_000815_070 |
0.1194032768 |
- |
- |
PREDICTED: methyltransferase-like protein 7A [Jatropha curcas] |
| 10 |
Hb_001123_040 |
0.1198182956 |
- |
- |
PREDICTED: R3H domain-containing protein 1-like [Jatropha curcas] |
| 11 |
Hb_010098_050 |
0.1219316113 |
- |
- |
PREDICTED: putative deoxyribonuclease tatdn3-A [Jatropha curcas] |
| 12 |
Hb_000260_630 |
0.123000741 |
- |
- |
PREDICTED: altered inheritance rate of mitochondria protein 25 isoform X1 [Jatropha curcas] |
| 13 |
Hb_089995_010 |
0.1230234728 |
- |
- |
PREDICTED: phosphatidylinositol glycan anchor biosynthesis class U protein-like [Eucalyptus grandis] |
| 14 |
Hb_000402_160 |
0.1230875246 |
- |
- |
PREDICTED: ribose-phosphate pyrophosphokinase 4 isoform X1 [Jatropha curcas] |
| 15 |
Hb_002228_070 |
0.123517683 |
- |
- |
inosine triphosphate pyrophosphatase, putative [Ricinus communis] |
| 16 |
Hb_002028_150 |
0.1262653986 |
- |
- |
PREDICTED: hepatocyte growth factor-regulated tyrosine kinase substrate [Jatropha curcas] |
| 17 |
Hb_000212_330 |
0.1270310729 |
- |
- |
PREDICTED: biotin carboxyl carrier protein of acetyl-CoA carboxylase 1, chloroplastic isoform X1 [Jatropha curcas] |
| 18 |
Hb_005245_080 |
0.129616306 |
- |
- |
PREDICTED: uncharacterized protein LOC105643645 isoform X2 [Jatropha curcas] |
| 19 |
Hb_002686_170 |
0.129641362 |
- |
- |
PREDICTED: uncharacterized protein LOC105632645 isoform X1 [Jatropha curcas] |
| 20 |
Hb_004070_020 |
0.1296984211 |
- |
- |
rhythmically-expressed protein 2 protein, putative [Ricinus communis] |