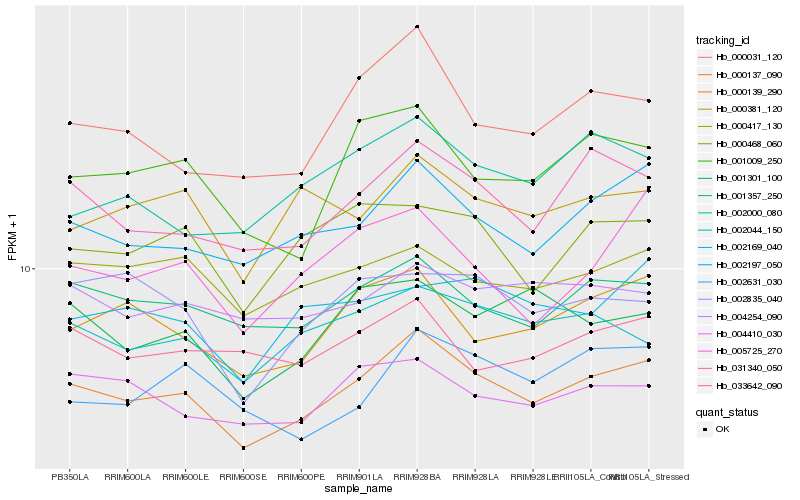
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000137_090 |
0.0 |
- |
- |
PREDICTED: HAUS augmin-like complex subunit 3 [Vitis vinifera] |
| 2 |
Hb_002197_050 |
0.0666656166 |
- |
- |
hypothetical protein CICLE_v10002157mg [Citrus clementina] |
| 3 |
Hb_031340_050 |
0.0699997896 |
- |
- |
PREDICTED: tropinone reductase 1-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_001357_250 |
0.0752628452 |
- |
- |
PREDICTED: UPF0505 protein C16orf62 homolog [Jatropha curcas] |
| 5 |
Hb_000031_120 |
0.0776148153 |
- |
- |
PREDICTED: uncharacterized protein LOC105637488 [Jatropha curcas] |
| 6 |
Hb_000139_290 |
0.0792994238 |
- |
- |
PREDICTED: peroxisomal adenine nucleotide carrier 1-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_001009_250 |
0.0813318873 |
- |
- |
PREDICTED: nucleoside diphosphate kinase 2, chloroplastic [Jatropha curcas] |
| 8 |
Hb_002044_150 |
0.0858726117 |
- |
- |
PREDICTED: A/G-specific adenine DNA glycosylase isoform X1 [Jatropha curcas] |
| 9 |
Hb_004410_030 |
0.0869359803 |
- |
- |
PREDICTED: protein ACCUMULATION AND REPLICATION OF CHLOROPLASTS 6, chloroplastic isoform X1 [Jatropha curcas] |
| 10 |
Hb_000468_060 |
0.0881454595 |
- |
- |
ku P70 DNA helicase, putative [Ricinus communis] |
| 11 |
Hb_000417_130 |
0.0890962476 |
- |
- |
PREDICTED: SAP30-binding protein isoform X2 [Jatropha curcas] |
| 12 |
Hb_000381_120 |
0.0912528965 |
- |
- |
PREDICTED: uncharacterized protein LOC105648175 [Jatropha curcas] |
| 13 |
Hb_001301_100 |
0.0921346582 |
- |
- |
transporter, putative [Ricinus communis] |
| 14 |
Hb_002000_080 |
0.0937675655 |
- |
- |
PREDICTED: polypyrimidine tract-binding protein homolog 3 isoform X2 [Jatropha curcas] |
| 15 |
Hb_005725_270 |
0.0941842644 |
- |
- |
PREDICTED: uncharacterized protein LOC105630802 isoform X1 [Jatropha curcas] |
| 16 |
Hb_002169_040 |
0.0942708715 |
- |
- |
unknown [Populus trichocarpa] |
| 17 |
Hb_033642_090 |
0.0948136099 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_004254_090 |
0.0948394869 |
- |
- |
PREDICTED: cleavage stimulating factor 64 [Jatropha curcas] |
| 19 |
Hb_002631_030 |
0.0950463872 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_002835_040 |
0.0957283092 |
- |
- |
PREDICTED: quinolinate synthase, chloroplastic [Jatropha curcas] |