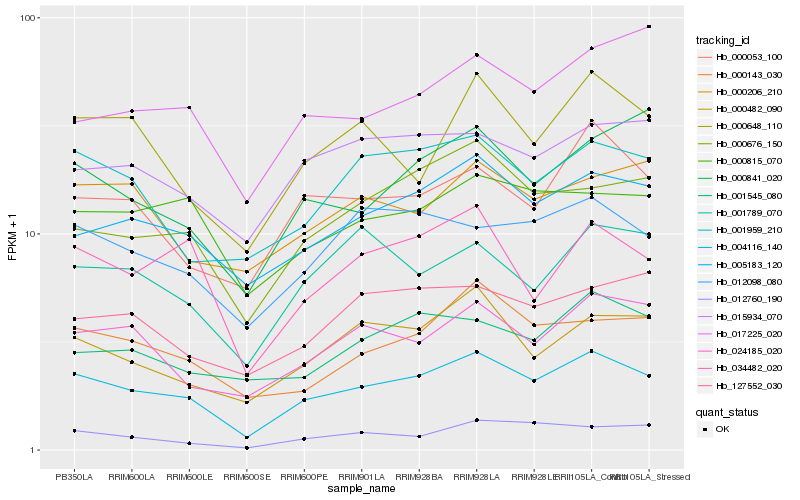
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004116_140 |
0.0 |
- |
- |
nuclear transcription factor, X-box binding, putative [Ricinus communis] |
| 2 |
Hb_005183_120 |
0.0954286597 |
- |
- |
protein translocase, putative [Ricinus communis] |
| 3 |
Hb_000143_030 |
0.1044120711 |
- |
- |
PREDICTED: uncharacterized protein LOC105629655 [Jatropha curcas] |
| 4 |
Hb_000676_150 |
0.1052724016 |
- |
- |
50S ribosomal protein L7/L12, putative [Ricinus communis] |
| 5 |
Hb_034482_020 |
0.1061472253 |
- |
- |
PREDICTED: bifunctional aspartokinase/homoserine dehydrogenase 1, chloroplastic-like [Jatropha curcas] |
| 6 |
Hb_000482_090 |
0.1072826427 |
- |
- |
PREDICTED: uncharacterized protein LOC105638145 [Jatropha curcas] |
| 7 |
Hb_024185_020 |
0.107497052 |
- |
- |
- |
| 8 |
Hb_012098_080 |
0.1083906951 |
- |
- |
KOM, putative [Ricinus communis] |
| 9 |
Hb_001959_210 |
0.1084632711 |
- |
- |
PREDICTED: uncharacterized protein LOC105638438 [Jatropha curcas] |
| 10 |
Hb_000053_100 |
0.1106520369 |
- |
- |
PREDICTED: uncharacterized protein LOC105642071 [Jatropha curcas] |
| 11 |
Hb_000648_110 |
0.1115397752 |
- |
- |
PREDICTED: D-xylose-proton symporter-like 3, chloroplastic [Jatropha curcas] |
| 12 |
Hb_000815_070 |
0.1126839739 |
- |
- |
PREDICTED: methyltransferase-like protein 7A [Jatropha curcas] |
| 13 |
Hb_000841_020 |
0.114383912 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 10-B-like [Populus euphratica] |
| 14 |
Hb_015934_070 |
0.1169887171 |
- |
- |
PREDICTED: ribose-phosphate pyrophosphokinase 4 isoform X1 [Jatropha curcas] |
| 15 |
Hb_127552_030 |
0.1202311225 |
- |
- |
PREDICTED: uncharacterized protein LOC105629655 [Jatropha curcas] |
| 16 |
Hb_001545_080 |
0.1206842287 |
- |
- |
PREDICTED: tRNA pseudouridine(38/39) synthase isoform X3 [Jatropha curcas] |
| 17 |
Hb_001789_070 |
0.1207003741 |
- |
- |
PREDICTED: probable prolyl 4-hydroxylase 12 [Jatropha curcas] |
| 18 |
Hb_012760_190 |
0.1213484912 |
- |
- |
PREDICTED: fatty acyl-CoA reductase 2 [Jatropha curcas] |
| 19 |
Hb_000206_210 |
0.1221247942 |
- |
- |
PREDICTED: G patch domain-containing protein 11 [Jatropha curcas] |
| 20 |
Hb_017225_020 |
0.127194895 |
- |
- |
PREDICTED: protein BRICK 1 [Gossypium raimondii] |