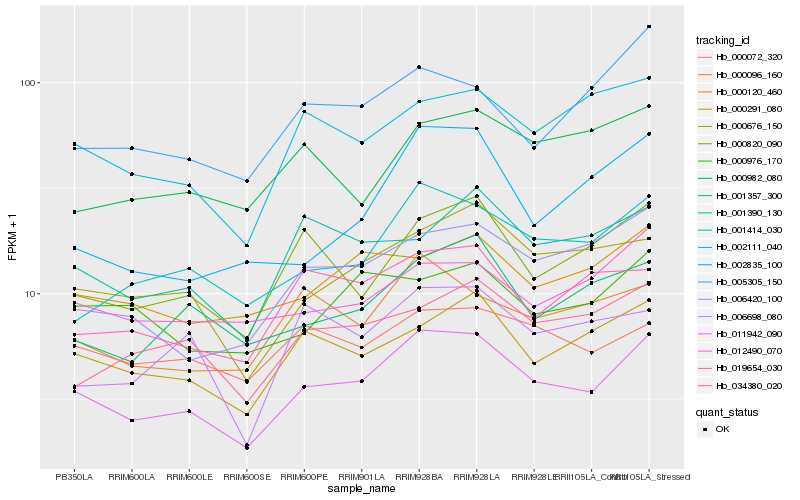
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011942_090 |
0.0 |
- |
- |
PREDICTED: serine/threonine-protein kinase OSR1 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000676_150 |
0.099878425 |
- |
- |
50S ribosomal protein L7/L12, putative [Ricinus communis] |
| 3 |
Hb_000291_080 |
0.1008609986 |
- |
- |
Vacuolar protein sorting-associated protein VPS9, putative [Ricinus communis] |
| 4 |
Hb_001414_030 |
0.1009510936 |
- |
- |
PREDICTED: aspartate-semialdehyde dehydrogenase [Jatropha curcas] |
| 5 |
Hb_000820_090 |
0.1041530436 |
- |
- |
- |
| 6 |
Hb_006420_100 |
0.1063533959 |
- |
- |
PREDICTED: protein bicaudal C homolog 1 [Jatropha curcas] |
| 7 |
Hb_002111_040 |
0.1108629064 |
- |
- |
hypothetical protein JCGZ_16989 [Jatropha curcas] |
| 8 |
Hb_001357_300 |
0.1113020617 |
- |
- |
PREDICTED: imidazole glycerol phosphate synthase hisHF, chloroplastic isoform X1 [Jatropha curcas] |
| 9 |
Hb_000096_160 |
0.1124128118 |
transcription factor |
TF Family: MYB-related |
PREDICTED: telomere repeat-binding factor 2 [Jatropha curcas] |
| 10 |
Hb_034380_020 |
0.1151924899 |
- |
- |
PREDICTED: phosphoglycerate mutase-like [Jatropha curcas] |
| 11 |
Hb_019654_030 |
0.1155331703 |
- |
- |
maoC-like dehydratase domain-containing family protein [Populus trichocarpa] |
| 12 |
Hb_000120_460 |
0.1175160884 |
- |
- |
PREDICTED: thioredoxin reductase NTRB-like [Jatropha curcas] |
| 13 |
Hb_001390_130 |
0.1189304871 |
- |
- |
hypothetical protein POPTR_0009s08590g [Populus trichocarpa] |
| 14 |
Hb_012490_070 |
0.1192238142 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000072_320 |
0.119580356 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RLIM-like [Jatropha curcas] |
| 16 |
Hb_005305_150 |
0.1228676399 |
- |
- |
PREDICTED: 40S ribosomal protein S2-4-like [Jatropha curcas] |
| 17 |
Hb_002835_100 |
0.1243253697 |
- |
- |
PREDICTED: protein DEHYDRATION-INDUCED 19 homolog 6-like [Jatropha curcas] |
| 18 |
Hb_000976_170 |
0.1247919862 |
- |
- |
PREDICTED: ATPase family AAA domain-containing protein 3-like [Jatropha curcas] |
| 19 |
Hb_006698_080 |
0.126625173 |
- |
- |
PREDICTED: tobamovirus multiplication protein 1 isoform X2 [Jatropha curcas] |
| 20 |
Hb_000982_080 |
0.1275947673 |
desease resistance |
Gene Name: AAA |
PREDICTED: 26S proteasome regulatory subunit 4 homolog A [Jatropha curcas] |