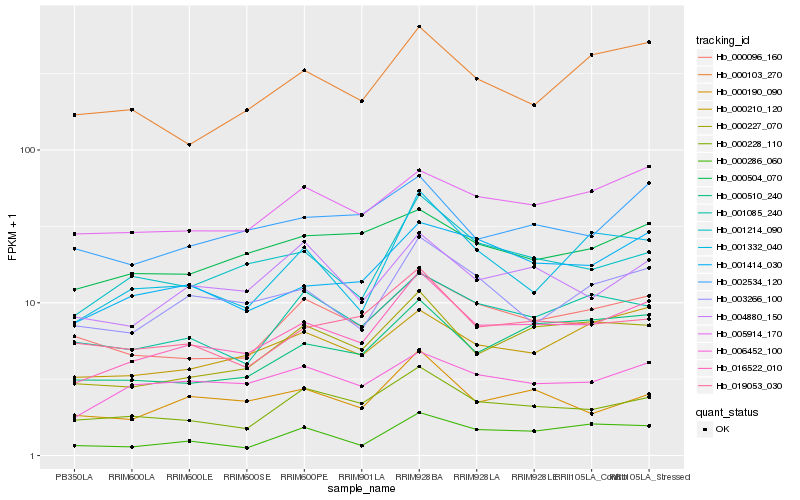
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_016522_010 |
0.0 |
- |
- |
PREDICTED: short-chain dehydrogenase/reductase 2b-like [Jatropha curcas] |
| 2 |
Hb_000227_070 |
0.0900542224 |
- |
- |
PREDICTED: ATP-dependent (S)-NAD(P)H-hydrate dehydratase isoform X2 [Jatropha curcas] |
| 3 |
Hb_001414_030 |
0.0949116749 |
- |
- |
PREDICTED: aspartate-semialdehyde dehydrogenase [Jatropha curcas] |
| 4 |
Hb_001214_090 |
0.0957350866 |
- |
- |
PREDICTED: acyl-coenzyme A oxidase 4, peroxisomal [Jatropha curcas] |
| 5 |
Hb_000510_240 |
0.1006065192 |
desease resistance |
Gene Name: PRK |
ATP binding protein, putative [Ricinus communis] |
| 6 |
Hb_000228_110 |
0.1028295608 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 7 |
Hb_000210_120 |
0.1033236289 |
- |
- |
PREDICTED: uncharacterized protein LOC105635829 isoform X1 [Jatropha curcas] |
| 8 |
Hb_003266_100 |
0.1041124602 |
- |
- |
Isomerase protein [Gossypium arboreum] |
| 9 |
Hb_000096_160 |
0.1044540843 |
transcription factor |
TF Family: MYB-related |
PREDICTED: telomere repeat-binding factor 2 [Jatropha curcas] |
| 10 |
Hb_005914_170 |
0.1049265514 |
- |
- |
PREDICTED: eukaryotic translation initiation factor isoform 4E-2-like [Gossypium raimondii] |
| 11 |
Hb_019053_030 |
0.1077654289 |
- |
- |
PREDICTED: sulfite oxidase [Jatropha curcas] |
| 12 |
Hb_006452_100 |
0.1100322473 |
transcription factor |
TF Family: Trihelix |
PREDICTED: uncharacterized protein LOC105629120 [Jatropha curcas] |
| 13 |
Hb_000103_270 |
0.1103967899 |
- |
- |
PREDICTED: 1,2-dihydroxy-3-keto-5-methylthiopentene dioxygenase 2 [Fragaria vesca subsp. vesca] |
| 14 |
Hb_002534_120 |
0.1114675894 |
- |
- |
PREDICTED: protein DCL, chloroplastic [Jatropha curcas] |
| 15 |
Hb_001085_240 |
0.1127525529 |
- |
- |
PREDICTED: probable calcium-binding protein CML22 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000504_070 |
0.1138067258 |
- |
- |
PREDICTED: pescadillo homolog isoform X1 [Jatropha curcas] |
| 17 |
Hb_000190_090 |
0.114290803 |
- |
- |
PREDICTED: uncharacterized protein LOC105649930 [Jatropha curcas] |
| 18 |
Hb_004880_150 |
0.1145084423 |
- |
- |
PREDICTED: uncharacterized protein LOC105632834 [Jatropha curcas] |
| 19 |
Hb_001332_040 |
0.1145456202 |
- |
- |
PREDICTED: transcription initiation factor IIB [Fragaria vesca subsp. vesca] |
| 20 |
Hb_000286_060 |
0.1156804494 |
- |
- |
PREDICTED: uncharacterized protein LOC105633357 [Jatropha curcas] |