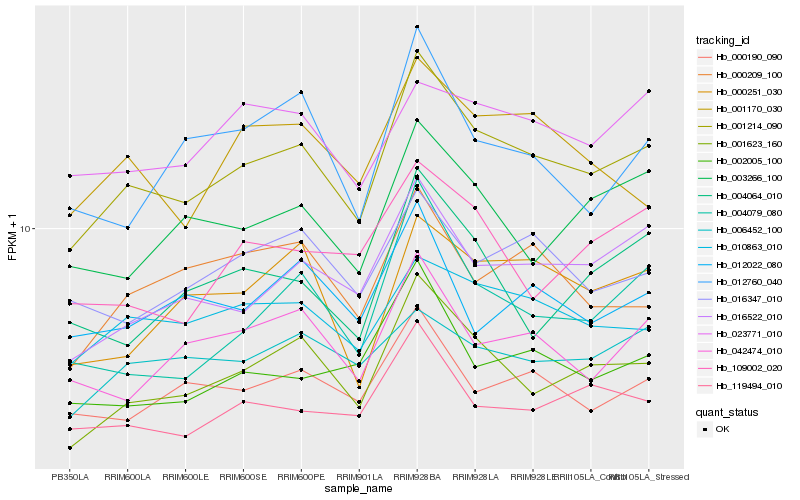
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001214_090 |
0.0 |
- |
- |
PREDICTED: acyl-coenzyme A oxidase 4, peroxisomal [Jatropha curcas] |
| 2 |
Hb_001623_160 |
0.0943478042 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At2g33170 [Jatropha curcas] |
| 3 |
Hb_016522_010 |
0.0957350866 |
- |
- |
PREDICTED: short-chain dehydrogenase/reductase 2b-like [Jatropha curcas] |
| 4 |
Hb_010863_010 |
0.1027276634 |
transcription factor |
TF Family: C3H |
nucleic acid binding protein, putative [Ricinus communis] |
| 5 |
Hb_000190_090 |
0.1055829945 |
- |
- |
PREDICTED: uncharacterized protein LOC105649930 [Jatropha curcas] |
| 6 |
Hb_119494_010 |
0.1097823796 |
- |
- |
PREDICTED: box C/D snoRNA protein 1 [Jatropha curcas] |
| 7 |
Hb_003266_100 |
0.1108020237 |
- |
- |
Isomerase protein [Gossypium arboreum] |
| 8 |
Hb_042474_010 |
0.1113330017 |
- |
- |
PREDICTED: adenylyltransferase and sulfurtransferase MOCS3 [Jatropha curcas] |
| 9 |
Hb_016347_010 |
0.1129619555 |
- |
- |
protein transporter, putative [Ricinus communis] |
| 10 |
Hb_001170_030 |
0.1140379739 |
- |
- |
synaptonemal complex protein, putative [Ricinus communis] |
| 11 |
Hb_002005_100 |
0.1151099519 |
- |
- |
- |
| 12 |
Hb_004079_080 |
0.1161418222 |
- |
- |
PREDICTED: ras-related protein RHN1-like [Jatropha curcas] |
| 13 |
Hb_000251_030 |
0.118186673 |
- |
- |
nicotinate phosphoribosyltransferase, putative [Ricinus communis] |
| 14 |
Hb_000209_100 |
0.1187599861 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 2 isoform X2 [Jatropha curcas] |
| 15 |
Hb_006452_100 |
0.1194005372 |
transcription factor |
TF Family: Trihelix |
PREDICTED: uncharacterized protein LOC105629120 [Jatropha curcas] |
| 16 |
Hb_012760_040 |
0.1209201515 |
- |
- |
PREDICTED: aconitate hydratase 1 [Jatropha curcas] |
| 17 |
Hb_004064_010 |
0.1215030107 |
- |
- |
PREDICTED: uncharacterized protein LOC105133340 isoform X1 [Populus euphratica] |
| 18 |
Hb_012022_080 |
0.1215288958 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 19 |
Hb_109002_020 |
0.1243323206 |
- |
- |
PREDICTED: CTP synthase-like [Jatropha curcas] |
| 20 |
Hb_023771_010 |
0.1251444495 |
- |
- |
basic helix-loop-helix-containing protein, putative [Ricinus communis] |