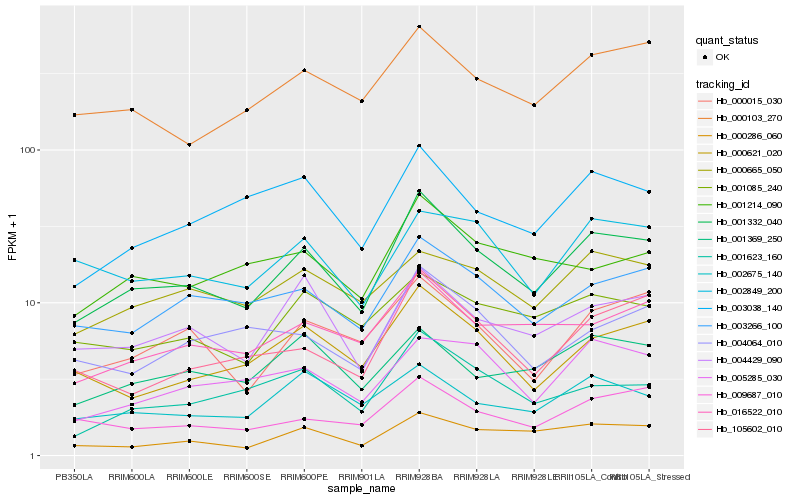
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001332_040 |
0.0 |
- |
- |
PREDICTED: transcription initiation factor IIB [Fragaria vesca subsp. vesca] |
| 2 |
Hb_000286_060 |
0.1081764163 |
- |
- |
PREDICTED: uncharacterized protein LOC105633357 [Jatropha curcas] |
| 3 |
Hb_003266_100 |
0.109011945 |
- |
- |
Isomerase protein [Gossypium arboreum] |
| 4 |
Hb_016522_010 |
0.1145456202 |
- |
- |
PREDICTED: short-chain dehydrogenase/reductase 2b-like [Jatropha curcas] |
| 5 |
Hb_000103_270 |
0.1155049515 |
- |
- |
PREDICTED: 1,2-dihydroxy-3-keto-5-methylthiopentene dioxygenase 2 [Fragaria vesca subsp. vesca] |
| 6 |
Hb_000621_020 |
0.1156176253 |
- |
- |
PREDICTED: mannose-6-phosphate isomerase 1 [Jatropha curcas] |
| 7 |
Hb_004429_090 |
0.1158393879 |
- |
- |
PREDICTED: ras GTPase-activating protein-binding protein 1-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_000015_030 |
0.1177095137 |
- |
- |
PREDICTED: THO complex subunit 4B-like [Jatropha curcas] |
| 9 |
Hb_003038_140 |
0.1193867198 |
- |
- |
soluble inorganic pyrophosphatase [Hevea brasiliensis] |
| 10 |
Hb_001623_160 |
0.1194990671 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At2g33170 [Jatropha curcas] |
| 11 |
Hb_105602_010 |
0.1205057961 |
- |
- |
Ketose-bisphosphate aldolase class-II family protein isoform 5 [Theobroma cacao] |
| 12 |
Hb_009687_010 |
0.1231024804 |
- |
- |
PREDICTED: protein SUPPRESSOR OF K(+) TRANSPORT GROWTH DEFECT 1-like isoform X4 [Jatropha curcas] |
| 13 |
Hb_001085_240 |
0.1294336163 |
- |
- |
PREDICTED: probable calcium-binding protein CML22 isoform X1 [Jatropha curcas] |
| 14 |
Hb_005285_030 |
0.1305657876 |
- |
- |
hypothetical protein JCGZ_20905 [Jatropha curcas] |
| 15 |
Hb_002675_140 |
0.1314323711 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase At1g63170 [Jatropha curcas] |
| 16 |
Hb_004064_010 |
0.1320988618 |
- |
- |
PREDICTED: uncharacterized protein LOC105133340 isoform X1 [Populus euphratica] |
| 17 |
Hb_000665_050 |
0.1321461882 |
- |
- |
PREDICTED: aberrant root formation protein 4 [Jatropha curcas] |
| 18 |
Hb_002849_200 |
0.1321684496 |
- |
- |
hypothetical protein JCGZ_15712 [Jatropha curcas] |
| 19 |
Hb_001214_090 |
0.1323344447 |
- |
- |
PREDICTED: acyl-coenzyme A oxidase 4, peroxisomal [Jatropha curcas] |
| 20 |
Hb_001369_250 |
0.1324244376 |
- |
- |
PREDICTED: uncharacterized protein LOC105646469 isoform X1 [Jatropha curcas] |